Abstract
In this blog, we analyze the non-deep learning components of DeepVariant in detail. We suggest that innovations in how DeepVariant represents allele candidates are key to its ability to solve hard cases, for example complex multi-allelic sites. These insights may prove useful both in developing better deep learning methods, and in improving non-deep learning methods that operate on genomic data.
Introduction
Variant calling is the fundamental process of realizing the power of human genome sequencing to gain insights for precision medicine. At the birth of next-generation sequencing (NGS) about 15 years ago, if one sequenced the same individual twice, the concordance between the two independent variant calling results could be quite poor. Researchers have reported the concordance between different variant calling pipelines to be merely at 60% as recently as 2013. Now, with 5 or 6 more years of research and development work, variant calling has become highly reproducible – and researchers are looking for perfection.
More recently, the Genome In A Bottle (GIAB) consortium, led by Justin Zook and Marc Salit from NIST, has developed standard material and variant call sets for setting a gold standard to help the genomics community benchmarking variant call methods. These materials were essential to the PrecisionFDA challenges conducted by DNAnexus on behalf of the US FDA. Pioneered by Ryan Poplin and Mark DePristo from Google Brain, DeepVariant utilizes the GIAB call sets to train a deep learning model (InceptionNet v3) to solve the variant calling problem. DeepVariant is distinguished by its high accuracy, illustrated in the DeepVariant paper and in multiple independent benchmarks (for example in PrecisionFDA, DNAnexus and recent papers). More importantly, it can be easily re-trained for different platforms, e.g. BGI-SEQ, without manually fine-tuning the model parameters to get better results, as long as a training set can be built by sequencing the samples from GIAB.
While the artificial neural network seems to magically solve some difficult problems, pre-processing and post-processing are critical for the final outcome. In DeepVariant or any general variant callers, the processing steps involve taking the alignment reads, high-confident variant call set, and reference sequences to generate tensors with correct labels for training.
Google Brain team has developed a library, called Nucleus, for bringing bioinformatics data files into TensorFlow records through Google’s Protocol Buffers. The Nucleus library takes care of the “extract, transform and load” (ETL) part for working with genomic data. We encourage anyone interested in processing genomics data to take a look at the library, as it can be very useful for the development of new variant calling algorithms or data processing pipelines.
Beyond ETL, we believe that correctly labeling complicated variant calling cases is essential for the success of DeepVariant. We are grateful that Google Brain released DeepVariant as an open-source project – this allows us to dive into the DeepVariant source code and perform experiments. This “Deep Dive into DeepVariant” is about our understanding of (1) the upstream bioinformatics processes for labeling and creating a training set for the deep neural network, and (2) the downstream processes for generating the final variant calls. While this blog and associated Jupyter notebooks (for case 1, 2, and 3) are specific to DeepVariant, we think these upstream and downstream processes can be generalized to improve both deep learning and non-deep learning methods. Here we hope to provide a bit more insight into DeepVariant for those interested in deep learning and the art of variant calling.
A Variant Candidate with Multiple Alternative Alleles
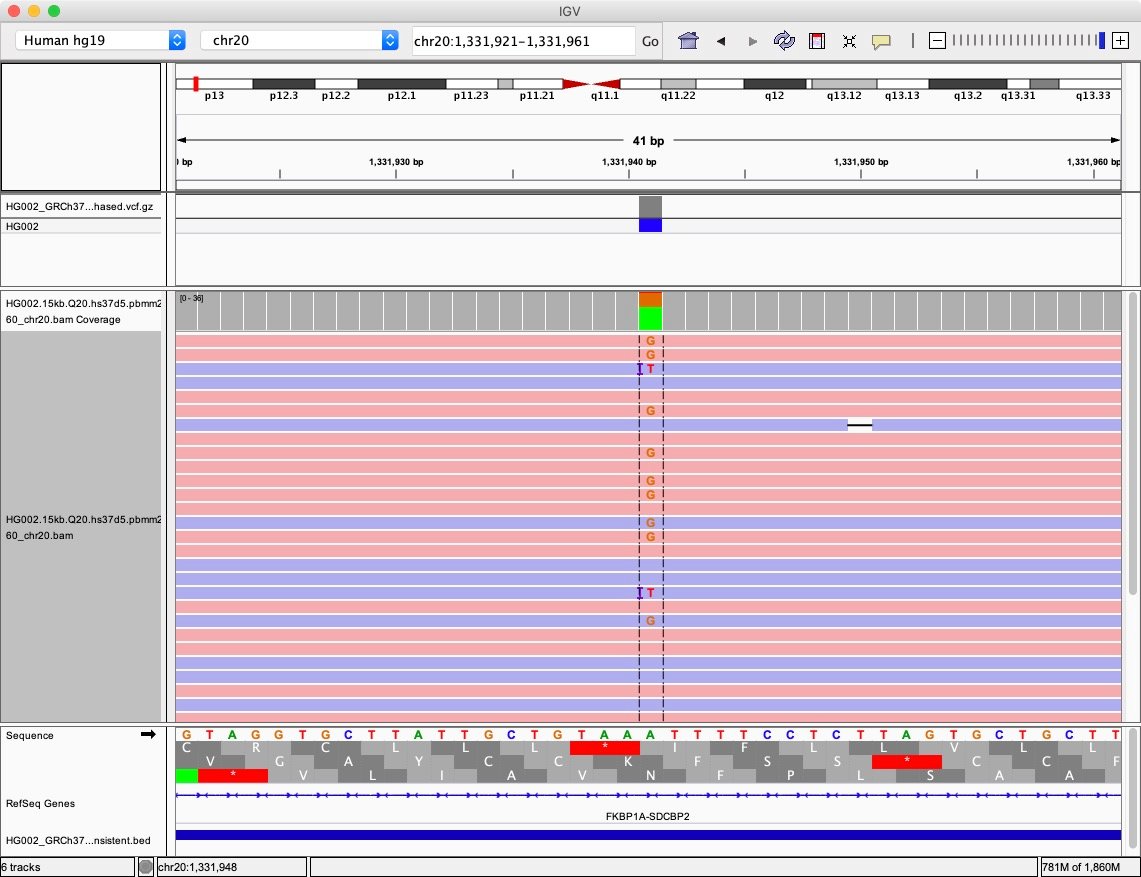
The Integrated Genome Viewer (IGV) snapshot below (Fig. 1) shows an interesting case for variant calling. In the central column, the reference allele is ‘A’, but there is more than one alternative allele: ‘G’ and ‘T’. We know that the true genotype here is ‘A/G’ here. How does DeepVariant generate the tensors (which are equivalent to multi-channel images) from the alignments for training?
In this case, the ‘T’ allele is likely to be an error. DeepVariant can filter out an allele if its frequency is lower than a certain threshold to avoid calling an erroneous allele. However, it will also be intriguing to lower the ` –vsc_min_fraction_snps` option from the default value 0.12 to 0.05 to incorporate the two alternative alleles ‘G’ and ‘T’ and see how DeepVariant reacts.
How Does DeepVariant Label Such Training Examples?
First, we’d like to know what kind of labels are generated inside DeepVariant. As a machine learning classifier, DeepVariant uses these labels to make examples for classification. The exact same process of making examples is used both in training DeepVariant and in separate processes that make calls.
Given a VCF file that contains a high-quality curated variant set, DeepVariant makes “examples” by (1) generating candidates by parsing the CIGAR strings in a BAM field to collect potential variant candidates, and (2) comparing the candidate variants to the high confidence variant sets to label examples. For each example which consists of a tensor and a label used for deep learning training, there are only three possible labels 0, 1, and 2 for a diploid genome. The label indicates the multiplicity of the corresponding variant allele combinations.
The labeling schema can be a bit confusing, so here is an example. The function deepvariant.variant_caller.VariantCaller() will call a candidate with the reference allele as ‘A’, and two alternative alleles ‘G’ and ‘T’ at the locus 20:1331941. Since there is more than one allele, DeepVariant actually makes 3 examples that consider ‘G’, ‘T’, and ‘G/T’ as the possible alternative variant combinations. In this case, the ground truth is that the ‘G’ allele is the real heterozygous variant and the label for the example that considers ‘G’ as the right alternative allele should be 1. As the ‘T’ allele is likely from sequencing error and it is not the real heterozygous variant, the label for ‘T’ should be 0. DeepVariant also considers the third example that both ‘G’ and ‘T’ are both alternative variants. Since there is still only one correct haplotype allele ‘G’ in the set of {‘G’, ‘T’}, it will label such example as 1. In this case, the labeling table is summarized below:
Ground Truth: Ref: 'A', Het-alt: 'G' Example 0: locus: 20:1331941-1331941 reference base(s): A alternative base(s): ['G', 'T'] alt allele(s): ['G'] label: 1 Example 1: locus: 20:1331941-1331941 reference base(s): A alternative base(s): ['G', 'T'] alt allele(s): ['T'] label: 0 Example 2: locus: 20:1331941-1331941 reference base(s): A alternative base(s): ['G', 'T'] alt allele(s): ['G', 'T'] label: 1

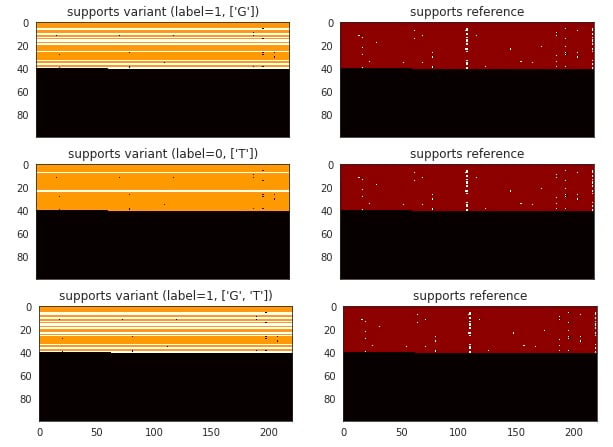
For the case shown in Fig. 1, DeepVariant generates three examples (Fig. 2 and Fig. 3). As the ‘T’ allele is most likely from sequencing error, the channel “support variant” only indicates there are three reads supporting ‘T’ allele and we want the ImageNet to ignore such errors by training it to classify such image to be labeled as “0”.
Such labeled examples are used for training the InceptionNet v3 model for an updated model. With the updated model, we can take unseen examples to generate the genotype probabilities that are consolidated by the post-processing step in DeepVariant for the final variant call output.
Post-Processing Genotype Probabilities for the Final Variant Call
The DeepVariant command `call_variants` will take unlabeled examples and generate the genotype probabilities for each example using InceptionNet as a classifier. For the examples at locus 20:1331941-1331941, with the default model from DeepVariant 0.7.2, the generated genotype probabilities are shown below:
Example 0 Call Variant Output: locus: 20:1331940-1331941 reference base(s): A alternative bases: ['G', 'T'] alt allele(s): ['G'] genotype probabilities: [0.0002641702, 0.9997351766, 6.532e-07] Example 1 Call Variant Output: locus: 20:1331940-1331941 reference base(s): A alternative bases: ['G', 'T'] alt allele(s): ['T'] genotype probabilities: [0.9992206097, 0.0007750907, 4.2996e-06] Example 2 Call Variant Output: locus: 20:1331940-1331941 reference base(s): A alternative bases: ['G', 'T'] alt allele(s): ['G', 'T'] genotype probabilities: [0.0001638031, 0.9998347759, 1.421e-06]
Here is one way to interpret this result: (1) when we consider the alternative allele ‘G’, there is a high probability there is one copy of that in the genome; (2) when we consider the alternative allele ‘T’, there is a very low probability (~ 0.0078 + 4.30e-06) that it is a real alternative allele; (3) when we consider both ‘G’ and ‘T’ are alternative alleles at the same time, there is a high probability (~0.9998) that we have one copy of ‘G’ or ‘T’. However, because ‘T’ is ruled out as a real allele, it also rules out that both ‘G’ and ‘T’ can be the alternative alleles. Thus, the final genotype should be ‘A/G’ with a reference base ‘A’ and a heterozygous variant ‘G’.
If you are interested in how the exact logic is performed by DeepVariant, you might want to take a look at the functions “merge_predictions()”, “get_alt_alleles_to_remove() “, “convert_call_variants_
What If There Are Indeed Two Alternative Alleles?
While it is not common that there are two alternative alleles simultaneously, there is still a small percentage of heterozygous loci where both variants are different from the reference base. We can consider one of these more complicated cases at the locus at chr20:4776366-4776367. The IGV screenshot shown below shows that the reference base is ‘G’, but the genotype is ‘T/C’.
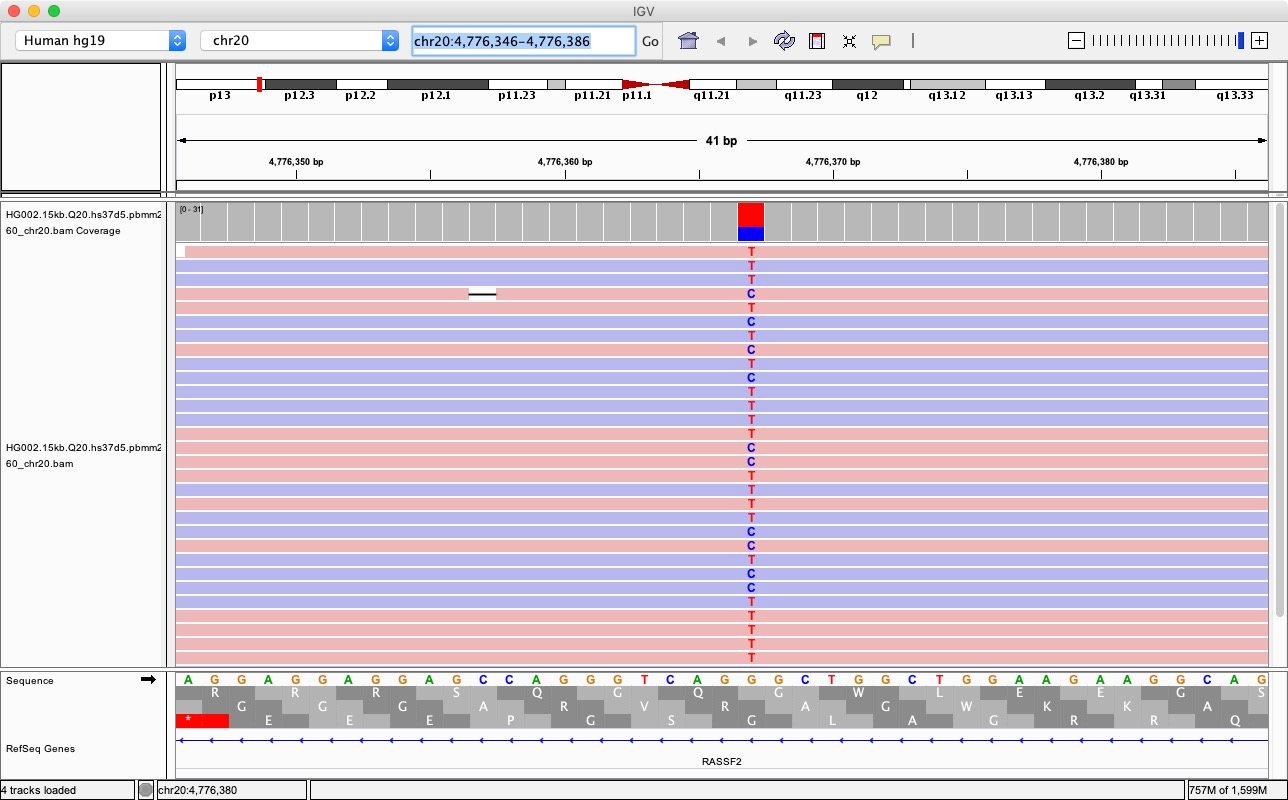
We apply call_variants to get the genotype probabilities:
Example 0 Call Variant Output: locus: 20:4776366-4776367 reference base(s): G alternative bases: ['C', 'T'] alt allele(s): ['C'] genotype probabilities: [4.98524e-05, 0.9999500513, 9.63e-08] Example 1 Call Variant Output: locus: 20:4776366-4776367 reference base(s): G alternative bases: ['C', 'T'] alt allele(s): ['T'] genotype probabilities: [6.3542e-06, 0.9999862909, 7.3549e-06] Example 2 Call Variant Output: locus: 20:4776366-4776367 reference base(s): G alternative bases: ['C', 'T'] alt allele(s): ['C', 'T'] genotype probabilities: [6.802e-07, 5.119e-07, 0.9999988079]
We can see that, in this case, when the DeepVariant considers the alternative alleles to be both ‘C’ and ‘T’, it gets the highest genotype probability. If we consider ‘C’ or ‘T’ independently, they also have a high probability to be “1”. This is indeed consistent with the heterozygous genotype ‘C/T’.
A Nasty Case of Many Alternative Alleles Around Indels and Homopolymers
The prior examples have shown variant cases which are relatively easier. As a tool, DeepVariant distinguishes itself in accuracy in the hard examples. This is the difference between 99.97% accuracy and 99.8% accuracy. Getting hard cases right is important because in a genome with 3 billion positions, an error rate of 1:10,000 means many false or missing calls, which could result in missed diagnoses or genetic associations, or in many variants of unknown significance in a clinical report.
Let’s examine a more complicated case where there are multiple alternative alleles considered by DeepVariant. Such situations are very common around homopolymer regions. Many alternative alleles could be identified by the calls_from_allele_counter() function inside DeepVariant’s VariantCaller module. Fig. 5 shows a case where there are alternative alleles identified as ‘A’, ‘ATATTT’, and ‘ATATTT’, with the reference allele ‘AT’ at locus 20:9068075-9068077.
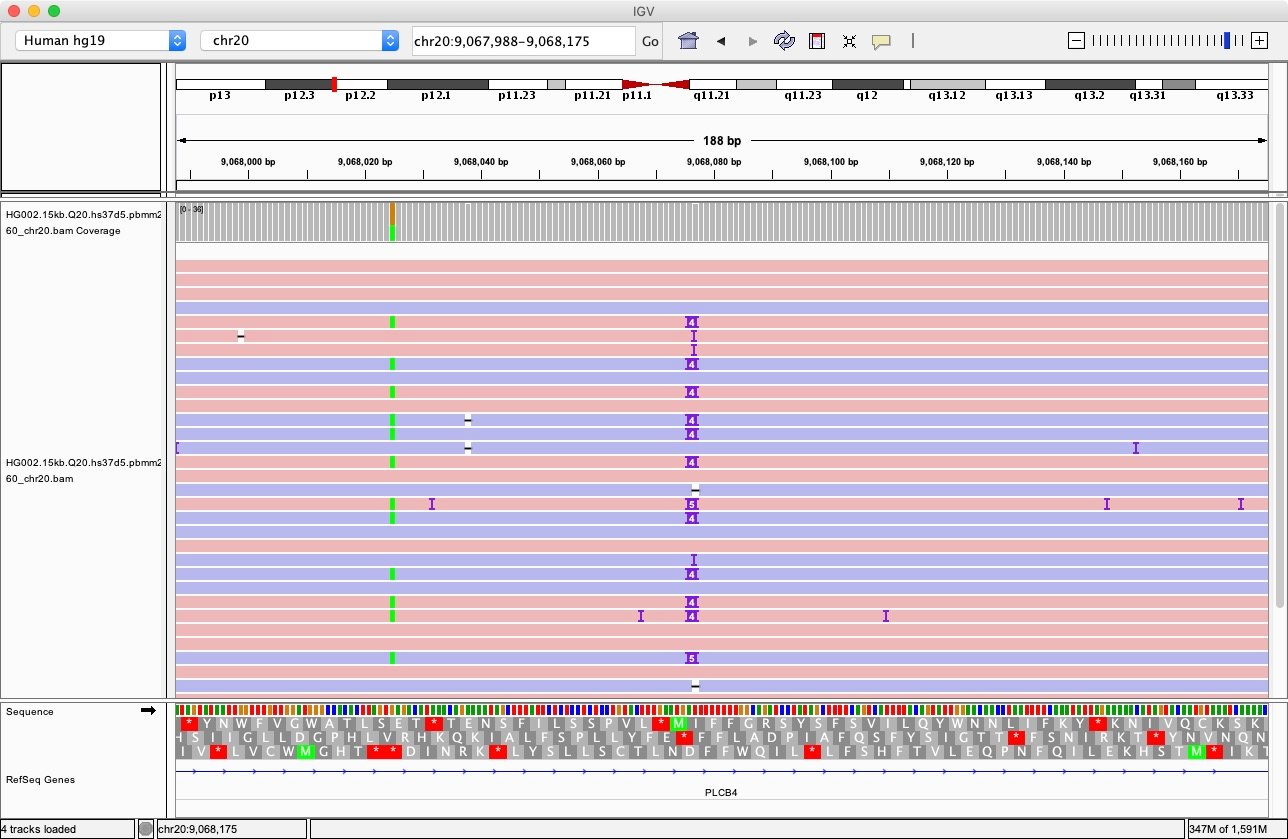
How many examples should DeepVariant generate for such a case? Including the reference allele, there are 4 possible alleles. DeepVariant generates examples of all possible combination of any of two different alleles; namely, the 6 combinations listed in Table 1.
| reference allele | alternative allele(s) | genotype label and probabilities | note | ||
| 0 | 1 | 2 | |||
| AT | A | 0.999 | 0.001 | 1.73E-06 | Reject ‘A’ as alt |
| AT | ATATTT | 0.271 | 0.729 | 1.86E-05 | ‘ATATTT’ is likely a het allele |
| AT | ATATTTT | 0.911 | 0.089 | 7.18E-05 | Reject ‘ATATTTT’ as alt |
| AT | A / ATATTT | 0.161 | 0.839 | 2.23E-05 | Consistent with ‘ATATTT’ as het |
| AT | A / ATATTTT | 0.929 | 0.071 | 2.37E-05 | Reject both ‘A’ and ‘ATATTTT’ |
| AT | ATATTT / ATATTTT | 0.002 | 0.998 | 1.16E-06 | Consistent with ‘ATATTT’ as het |
Table 1, The genotypes considered by DeepVariant and the probabilities inferred by its model. The highest genotype probabilities are highlighted in bold type for each example.
In Table 1, we highlight the highest genotype probability for each example. In this case, it is not hard to deduce to that the most likely alleles in this location are the reference allele and the ‘ATATTT’ alleles. Indeed, the true variant identified by the GIAB for this locus is a ‘TATT’ insertion, and this is consistent with a reference allele ‘AT’ and an alternative allele ‘ATATTT’ at this heterozygous site.
Final Thoughts
Here we examine a couple of cases of how DeepVariant labels variants for training, and how it aggregates the genotype probabilities returned from InceptionNet v3 for the final variant calls. Software like DeepVariant is complicated. This blog can only serve as a snapshot for us to better understand a small part of the codebase and the underlying algorithm. However, we hope that gaining insight on some of the details will help us to understand the strengths, and perhaps some weaknesses, so that we can apply the tool properly and resolve any future issues we encounter more efficiently.
After DeepVariant’s first preprint in 2016, more deep-learning based variant callers began to appear. For example, VariantNET and Clairvoyante were designed using multi-task learning for noisier raw single-molecule DNA sequencing reads. Clairvoyante trains a deep neural network to predict variant types (het or hom, SNP or indels) and alleles at the same time with a smaller network. In noisy long-read data, the make-examples step used by DeepVariant becomes very computationally expensive. DeepVariant may need to consider too many competing examples even for simple SNV cases because of the excessive errors in noisy reads. We think it makes aggregating the genotype probabilities correctly a harder problem. Clairvoyante generates only one tensor for each variant candidate. It could be beneficial for calling variants from noisy reads. In contrast, the more rigorous candidate generation and labeling process used in DeepVariant with a larger deep learning network may be essential for its best-in-class performance for high-quality data (e.g. read error rate < 1%).
The genomic community has come a long way, from scratching our collective heads to figure out why concordance was only at 60% just a couple years ago, to figure out what is going on for the residue errors after 99.9+% accuracy now. Along with the cheaper DNA sequencing and more versatile data types, deep learning-based variant callers such as DeepVariant will help scientists keep up with developing technology without significant re-investment in algorithm development. We are excited to see more applications using deep learning on genomic technologies to help us utilize genomic data like never before.
Acknowledgement
We thank Andrew Carroll and Pi-Chuan Chang from Google Brain for carefully reading the draft and giving suggestions for improvement. We also thank Samantha Zarate, Steve Osazuwa, Yih-Chii Hwang and Chai Fungtammasan from DNAnexus science team for fixing errors, typos and revising the draft thoroughly.


.png)
.png)
.png)