
While the amount and variety of genomics data available to scientists is greater now than ever before, large-scale bioinformatics studies can still seem daunting to researchers interested in exploring the applications of population genomics. Reducing the barrier to entry through software tools and open data access is essential to accelerate the path from genomic discovery to tailored patient care. In this light, we provide an introduction to a few fundamental types of genomic based studies.
Researchers leverage population-scale genomics in their studies to better understand how genetics contribute to an individual’s health. These large-scale studies start with the creation of cohorts — a group of individuals with a particular set of genotypes, phenotypes, and/or combination of the two — that the researcher is interested in studying. Traditional cohort creation for bioinformaticians is a process that involves using the command-line interface (CLI) and a variety of different genotype/phenotype specific tools to sort through datasets and parse hundreds of traits. DNAnexus Apollo enables researchers to work either within a JupyterLab/CLI environment or an intuitive user-interface to easily and quickly explore a multitude of phenotypes and genomic traits in the Cohort Browser, filter for desired traits, and visualize results using built-in charts and visualizations such as Manhattan plots.
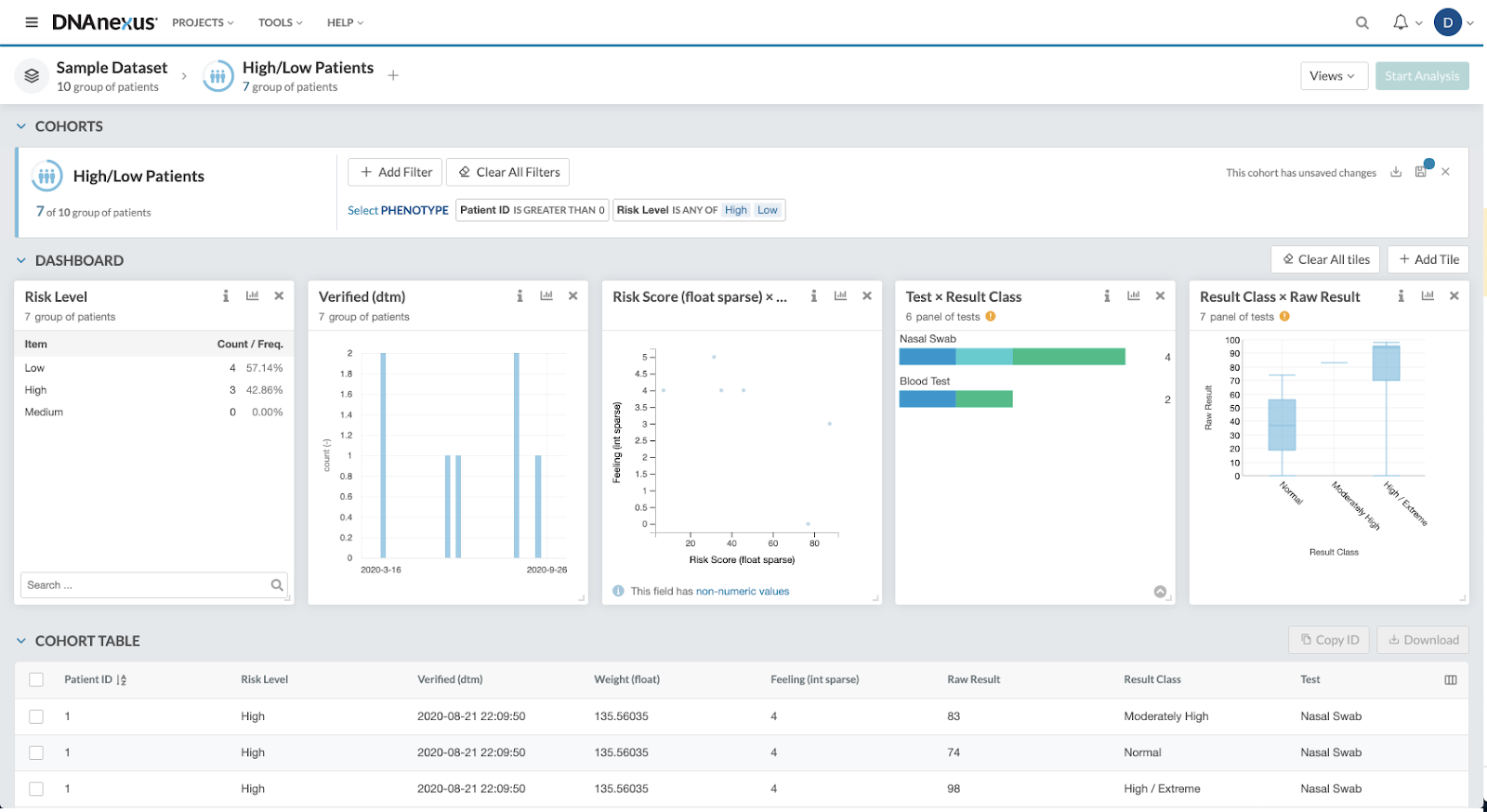
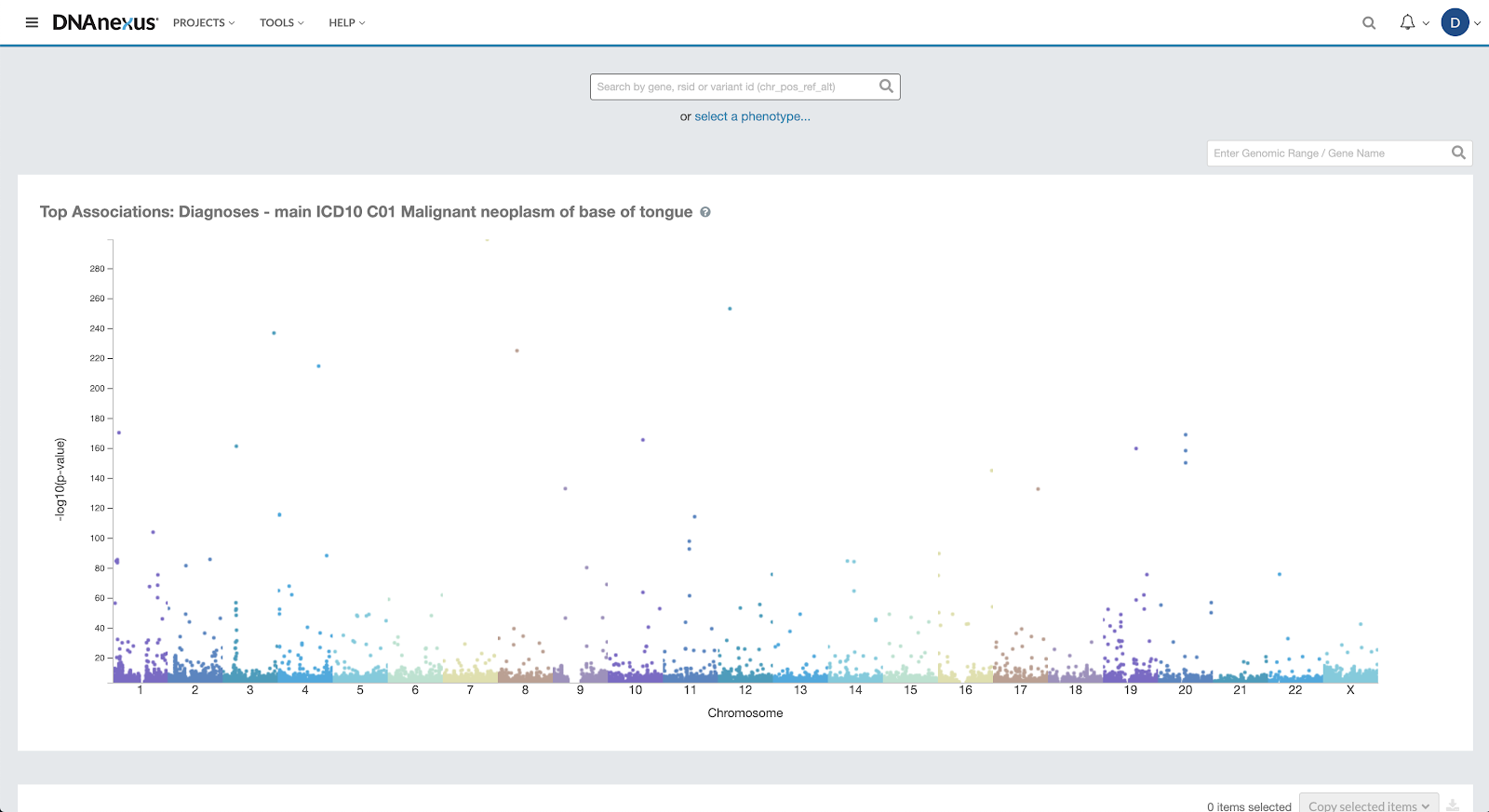
Once a cohort has been identified, two common analysis methods are genome- and phenome-wide association studies. A genome-wide association study (GWAS) is used to find associations between single-nucleotide polymorphisms (SNPs) and a certain trait or disease. Phenome-wide association studies (PheWAS) operate on a similar premise, and test genetic variants for associations with a set of phenotypes. Both types of studies provide information that researchers can use to uncover genetic risk factors for a variety of diseases and health conditions by surveying phenotype to SNP correlation. The insights gained into the disease associations provide a better understanding of underlying disease origins, and can aid in the discovery and validation of new targeted drug targets and/or preventative strategies.
SAIGE and PLATO are two common software algorithms that can be used to execute GWAS analysis. There are publicly available resources for how to conduct a GWAS using SAIGE, including this repository created by the Neale lab that details the results of a GWAS conducted on the UK Biobank dataset. Many of these guides provide specific details on how quality control was conducted, and which statistical analyses were used, so that those new to these types of studies can replicate their findings. PLATO offers features to those interested in analyzing phenotypes as well, and enables users to run both a GWAS or PheWAS using a single unified tool. Both of these algorithms are available on DNAnexus, wrapped in the scalable application framework accessible, allowing researchers to quickly create cohorts and analyze them in the same place.
Both of these types of studies will process large quantities of data when run on large populations, a challenge for most homegrown informatics systems. DNAnexus Apollo offers Jupyter notebooks that enable researchers to easily analyze the large scale data with actions like annotation of GWAS results, a critical step for making bioinformatics data more actionable for all researchers. Apollo is purpose-built to handle the scale and type of computations that population genomics studies use, allowing researchers to work with genomics and complex multi-omics data in exciting new ways.
More and more tools are becoming available that help enable researchers to conduct innovative and more involved analyses as well. Using the results from a GWAS, researchers can compare their variant sets with other databases online, and incorporate additional omics data types, such as RNA-seq, to better understand how variants affect gene expression. Or, by utilizing machine-learning, researchers can conduct fine-tuned analyses by combining GWAS and functional data to bring clarity to previously noisy results.
Population genomics studies hold the key to unlocking the potential of precision medicine. With tools like DNAnexus Apollo, researchers can more quickly utilize large and complex datasets for use in the identification of biological mechanisms of disease, the discovery and application of biomarkers, or omics-guided therapeutic target discovery.
Interested in learning more? Check out the recorded webinar with Ben Busby, Scientific Director, Research Platforms Outreach, and see how Apollo unlocks population-scale omics datasets for accelerated discovery.

.png)
.png)
.png)