
Everyone is happy to see 2020 receding in the rearview mirror. But while last year was difficult in so many respects, it was productive for the DNAnexus Product team. Here’s a list of some of the features and enhancements we released in 2020:
Improved Cohort Building and Analysis Experience
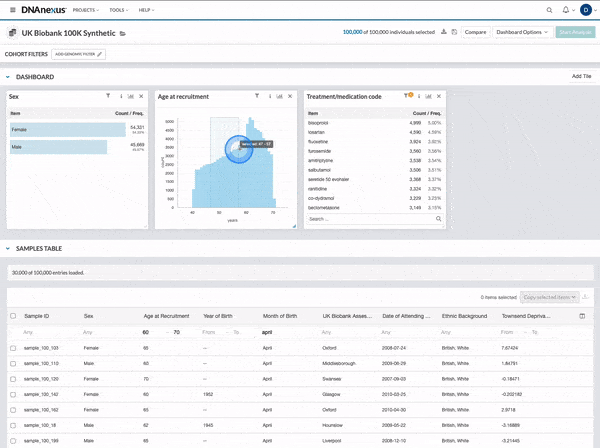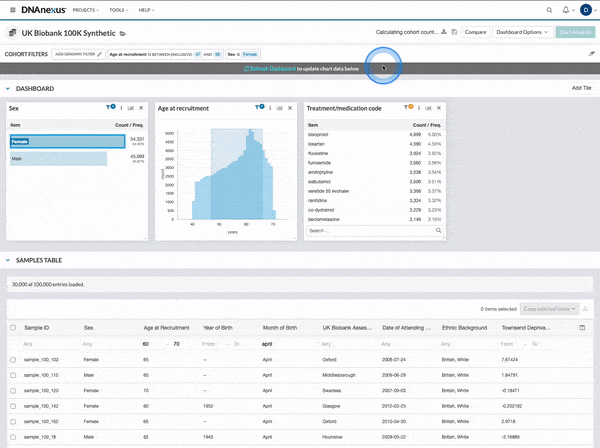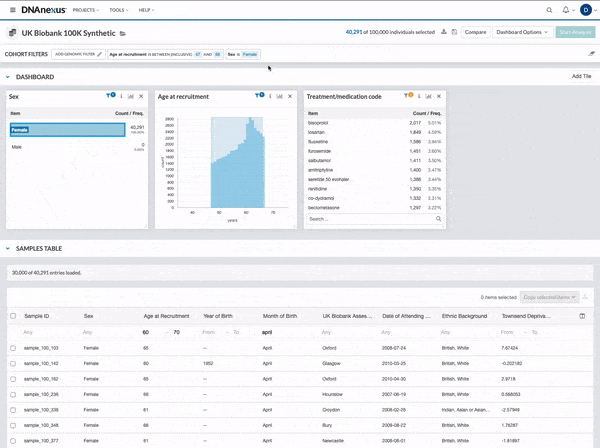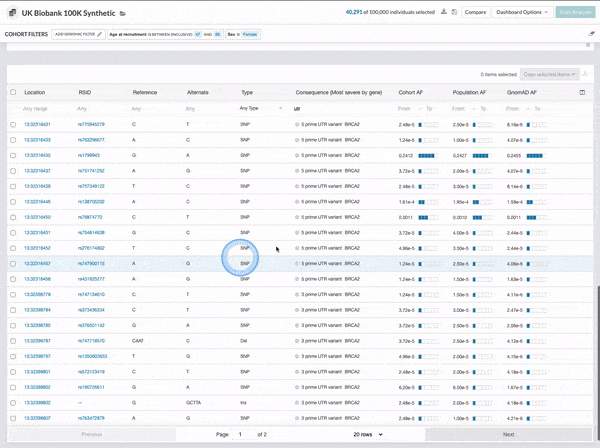
We made several updates to the Cohort Browser, to improve the experience of building and analyzing cohorts. The Cohort Browser now offers users:
- More options for customizing dashboards, through the use of custom filters and tiles, and Cohort Table field display options
- The ability to create and compare the phenotypic data for two cohorts, in support of use cases such as case-control comparisons, survival curve comparisons, and comparing a subpopulation to the whole
See our documentation for more on the improved Cohort Browser
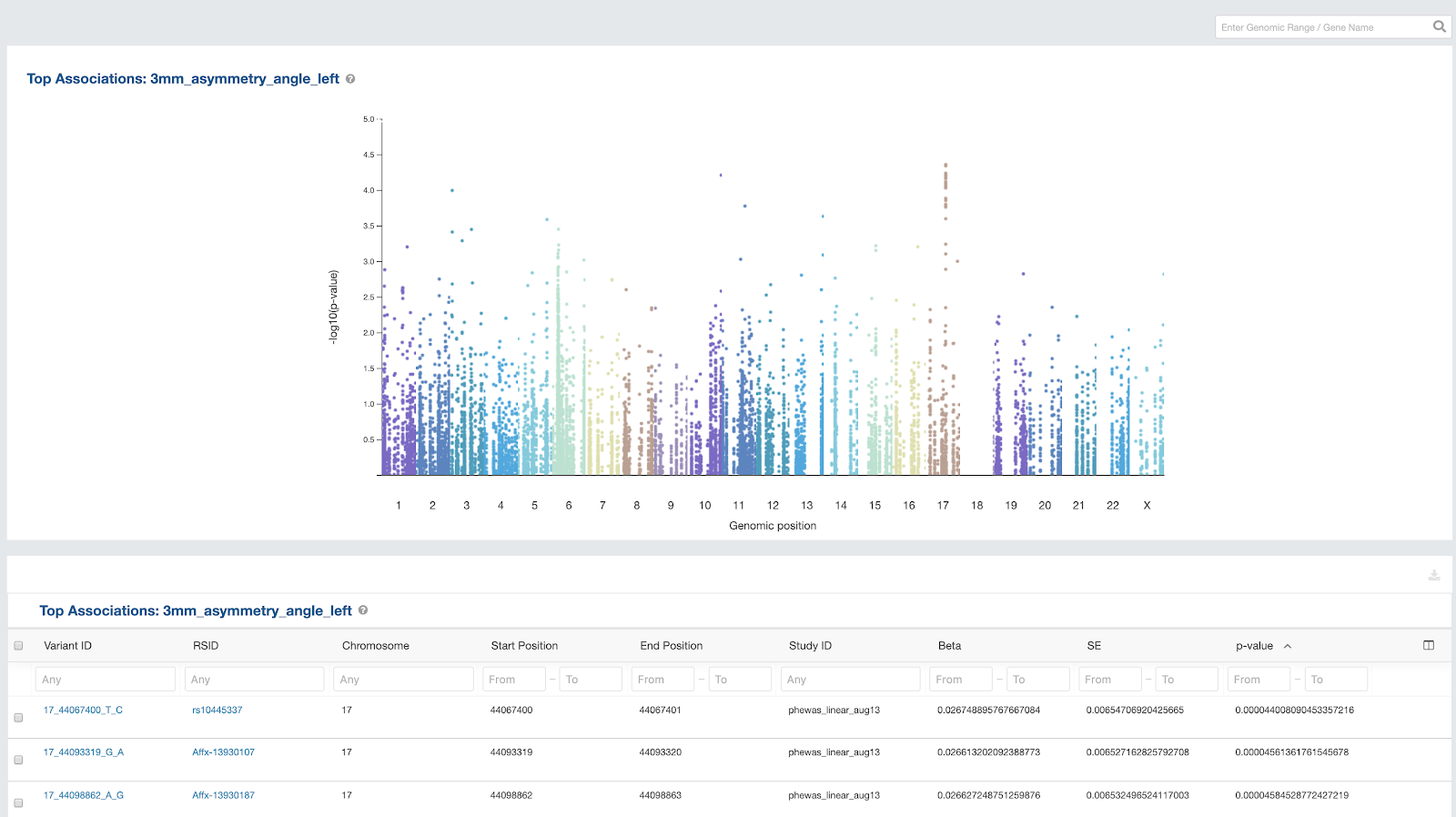
New Association Browser
Early in 2020, we released a new Association Browser, targeted to users conducting Genome Wide Association Studies (GWAS) and Phenome Wide Association Studies (PheWAS). The Association Browser enables these users to view and use an array of powerful visualizations – including zoomable Manhattan Plots – across a compendium of curated GWAS and PheWAS the results to more rapidly gain insight. See our documentation for more on the Association Browser.
Run GWAS Directly on Cohorts
Last summer, we made it possible for users quickly and easily to run PLATO and PLINK-based GWAS on cohorts built through the Cohort Browser, with additional configurations for case-control and specifying covariants. The results can then be viewed in an interactive Manhattan plot and variant table.
Data Ingestion Guidelines & Data Model Loader Documentation
In the fall, we published new documentation providing detailed guidance on how to ingest phenotypic data:
- Data ingestion overview
- Data Model Loader overview
- Guide to Data Model Loader support for different field types
- Guide to key steps in data ingestion
- Deep dive into Data Model Loader file inputs
- Data Model Loader walkthrough exercise
Extend Datasets with New/Derived Phenotype Information
The Dataset Extender app is an application meant to help expand your core dataset so that the entire team can access newly generated data. It is a lightweight app focused on quickly expanding core datasets with newly generated or acquired data that is to be shared with collaborators.
Running RStudio Shiny Server and Apps
With a few lines of R code, you can use RStudio Shiny to turn your command line apps and scripts into feature-rich, easy-to-use applications, accessible via a beautiful graphical user interface (GUI). This past summer, we published detailed guidelines on how to build and run RStudio Shiny apps on the DNAnexus Platform.
DX JupyterLab now supports papermill
DX JupyterLab now supports the use of papermill for automatically executing Jupyter notebooks and collecting the results. For more details, see the in-product DX JupyterLab documentation.
Migration to Python Version 3.0
The Python Software Foundation sunset Python version 2.x on January 1, 2020, and, moving forward, will support only Python 3.x. To learn more about how this affects the security of your Python apps and use of the DNAnexus Platform, please read the detailed FAQ we published last January.
Archive Service For Azure
For customers using Titan or Apollo on Azure, file-based archiving is now available in all Azure regions. Customers can take advantage of this feature to keep storage costs in check, by archiving files they use infrequently. Rest assured that archived files will be stored in accord with your organization’s data-retention policies, while maintaining security, file provenance tracking, and meta-data searchability. Learn more from this April 2020 post.
Leverage the Power of GPU Compute
Are you running ML- or AI-driven analyses that need the power of GPU compute? DNAnexus can help. DNAnexus now supports the use of all available AWS GPU instance types. Get the details from our documentation.
https Apps with Custom Hostnames
App developers can now provide access to their apps via user-friendly URLs. Each URL is generated by concatenating the app name with the name of the app host organization, e.g., https://myBestTool-acmeOrg.dnanexus.cloud/. Learn more by checking out our documentation.
dxWDL 2.0
dxWDL 2.0, the next generation of dxWDL, is on the way. Version 2.0 provides an improved WDL development experience, while following the WDL standard more closely, and removing dxWDL’s dependence on the Cromwell codebase. It relies on the DNAnexus wdlTools library for WDL parsing. If you’d like to check it out in advance of release, release candidate version 2.0.0-rc4 is available on Github.
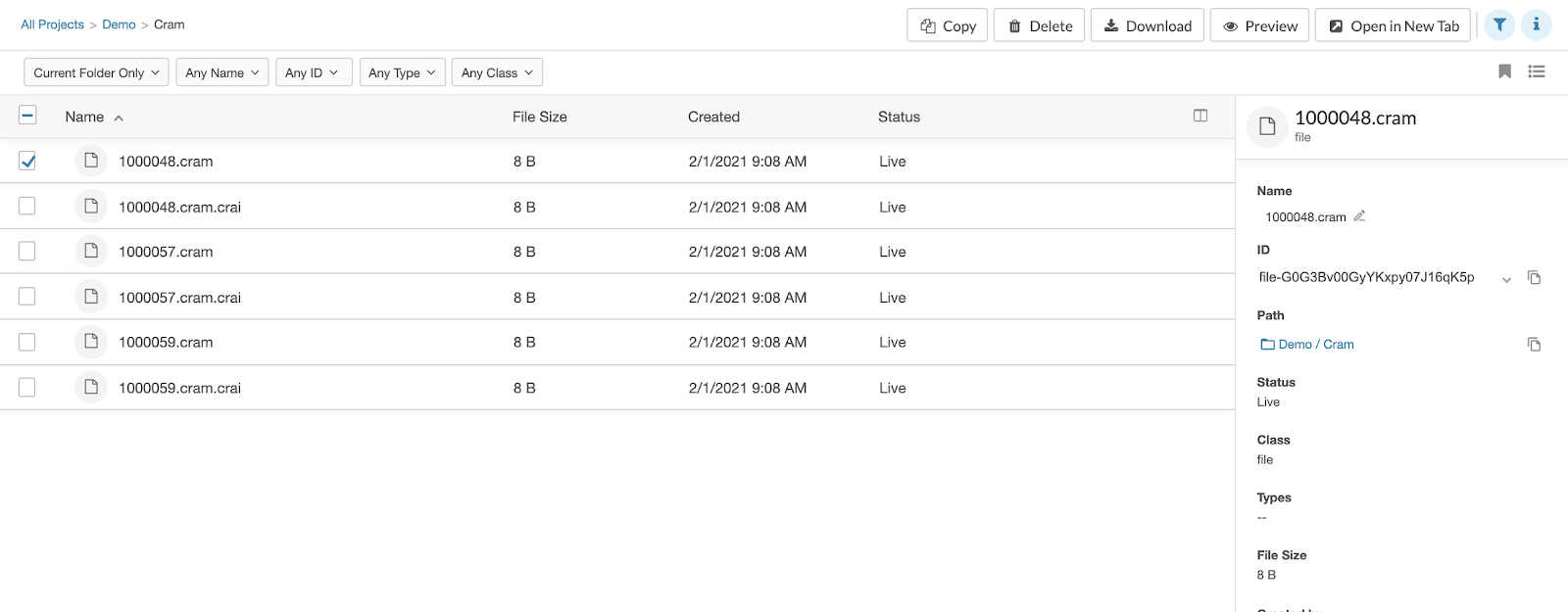
New File Info Pane
On enabling our new product UI, you’ll have access to a better way to get info on files. When viewing a list of files, click the “i” icon over the upper right corner of the list. Then tick the checkbox to the left of the name of the file about which you’d like to know more. Full file details will appear in the pane to the right of the list.

.png)
.png)
.png)