Authors:
The Broad Institute’s Genome Analysis Toolkit (GATK) is one of the most popular and well regarded repositories of best practices variant calling workflows, and DNAnexus has consistently provided optimized support of these pipelines on our platform. Announced January 9th, GATK4 is the latest release of the toolkit, and this release is particularly significant. GATK has been completely re-architected and is available fully open source. The best practices workflows descriptions are now also explicitly specified and distributed in an open format called the Workflow Description Language (WDL).
At DNAnexus, we are excited about supporting open, portable, and reproducible ways to share not only these new best practices workflows, but also general bioinformatics workflows written in WDL. As such, to execute explicit GATK4 workflow definitions written in WDL and maintained by the Broad, we use a new utility that we developed called dxWDL. With this tool, a GATK WDL workflow can be used just like any other workflow on the platform with all of the additional benefits our platform provides (e.g. provenance tracking, reproducibility, organization management, project collaboration, and security). As we did with GATK3, we are in the process of optimizing the performance of GATK4 on our platform and future posts will go into more detail about how it performs in terms of efficiency and accuracy. In the interim, we are pleased to announce the launch of the DNAnexus GATK4 Pilot Program, to be offered to a limited number of interested users, with broader access to the tool in the coming months. To request early access to GATK4 on DNAnexus please sign up here.
As an example of using GATK4 with dxWDL, we successfully ran a single sample haplotype workflow and Broad’s production germline variant calling workflow written in WDL on DNAnexus. For the production workflow, the version run on DNAnexus was modified slightly since Broad’s original pipeline has some Google cloud specific references. The following figure shows what the haplotype workflow looks like on DNAnexus: 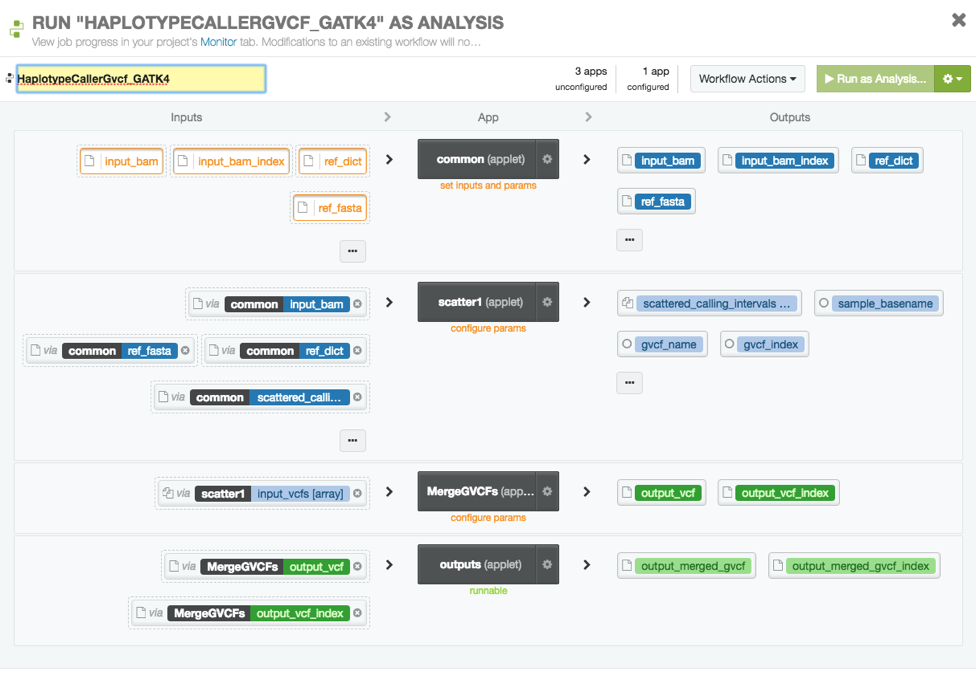
After execution, the timeline of tasks can be easily visualized showing Broad’s more complex production germline variant calling workflow:
Using dxWDL for GATK4 marks a change in how we will be executing these and other workflows written to be portable across platforms. In contrast to our previous approach of maintaining our own GATK applications, we will be directly supporting open and portable languages, such as WDL and CWL. Portability through languages such as WDL not only enables research in our field to be better critiqued and improved upon, but it also significantly reduces friction when communicating method details to collaborators and regulatory agencies. Often times while the details of a specific method in a workflow are not changed, some subtleties of in the workflow definition do change leading to reproducibility challenges, such as the changes we needed to make in the production pipeline described above. With our adoption of open workflow languages like WDL, we will more easily share these workflow-level differences with the community and work with one another towards a single representation that runs portably across a variety of execution platforms.
DNAnexus is proud to be one of the first genome informatics platforms to support WDL. As a member of the core team to govern future developments in WDL, we look forward to continuing work with the Broad and the broader community so that the the best practices WDL workflows can be run as efficiently and portably as possible.



.png)
.png)
.png)