
In the majority of cancer patients, the first sign that something is wrong is finding an already well-established tumor. But what if we could detect cancer at the first sign that the immune system is unable to correct it, when there are only a few cancer cells present? This is one of the challenging problems that TwinStrand Biosciences hopes to solve with their error correction technology, Duplex Sequencing™.
While current next generation sequencing (NGS) methods positively impact how we conduct research and discovery and stratify patients into disease categories, they aren’t yet sensitive enough to detect a small number of cancer cells among millions of healthy cells in the blood. Nor are they sensitive enough to detect the rare mutations induced by carcinogen exposure. And this is where the TwinStrand Duplex Sequencing Technology™ comes in.
The many steps involved in a typical NGS workflow — DNA isolation, fragmentation, amplification, etc. — can all introduce technical errors. Likewise, different sequencing platforms possess idiosyncratic differences that might also introduce errors. The goal of any NGS run and associated bioinformatic pipeline is to correct the error and separate the “noise” from true signal. However, TwinStrand takes error detection to a whole new level through use of their proprietary error correction technique that detects and masks technical errors with much greater sensitivity by examining and comparing both strands of a DNA sequence.
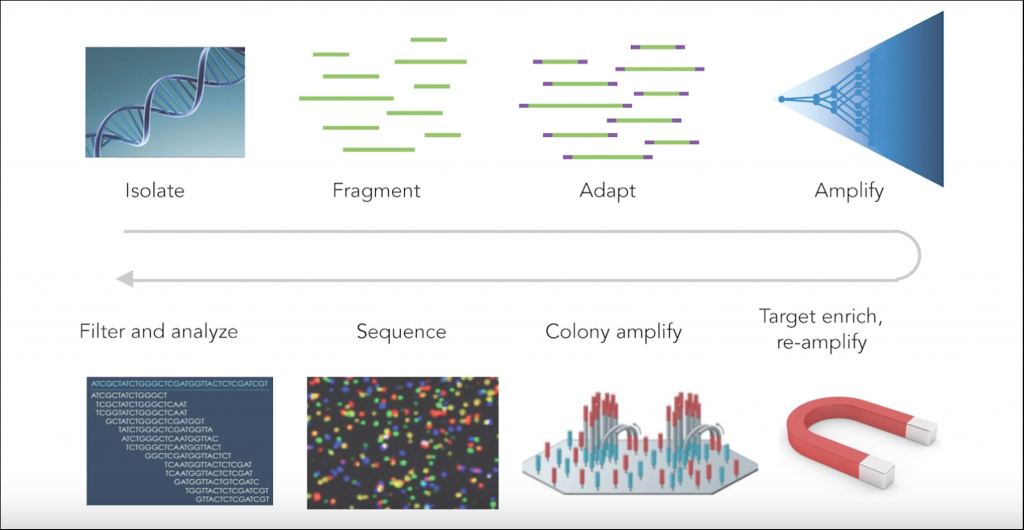
Typical sources of error in next generation sequencing (NGS)
Duplex Sequencing™ is an ultra-high accuracy sequencing method that overcomes the limitations of Next-Generation Sequencing by independently tracking both strands of individual DNA molecules. The paired sequences are compared to eliminate technical errors affecting only one strand, revealing ultra-low frequency biological mutation signal. Using a combination of proprietary biochemistry and cloud-based informatics, Duplex Sequencing greatly increases the resolution of NGS by reducing error rates from about 1-in-100 with standard NGS to below 1-in-10 million.
Duplex Sequencing can be used on any Illumina® sequencing platform. While early cancer detection and testing for residual disease make for natural applications of the technology, TwinStrand anticipates that the technology will be used to detect the emergence of antibiotic or antiviral resistance, occupational or environmental exposure to carcinogens, fetal abnormalities in non-invasive prenatal testing (NIPT), and in crime scene forensics.
Indeed, with the wide array of suitable applications, TwinStrand hopes to make their technology as accessible as possible — even to those with little NGS experience or bioinformatics expertise. One of the ways they’ve done this is by implementing their analytic pipeline on the DNAnexus Platform and making the platform available to their customers. When a sequencing run is complete, users can easily upload it to a DNAnexus TwinStrand Portal where the TwinStrand Biosciences analytical pipeline can be run. Then, users can access the results as well as share the data to collaborate with other users. Offering their pipeline on DNAnexus as part of their Duplex Sequencing assays made it easy for TwinStrand to deploy a complete kit-plus-informatics solution from the initial R&D on the pipeline through the final commercialization of the technology.
TwinStrand launched their technology with four new assays at the Advances in Genome Biology and Technology (AGBT) conference on Marco Island, FL, February 23-26. Specifically, they presented the use of their technology to measure residual disease in acute myeloid leukemia (AML MRD) and mutagenesis in human, mouse, and rat models. For more information contact info@twinstrandbio.com.

.png)
.png)
.png)