
When genetic tests are ordered, there’s probably little thought as to all of the bioinformatics work required to make the test possible. However, the bioinformatics team at Myriad Genetics understands firsthand just how much work it takes. Myriad Genetics provides diagnostic tests to help physicians understand risk profiles, diagnose medical conditions, or inform treatment decisions. To support their comprehensive test menu and commitment to providing timely and accurate test results, the bioinformatics team at Myriad focuses on optimizing their bioinformatics pipelines. How? By designing pipelines to leverage modularity and computational re-use to make improvements and iterate more quickly.
Jeffrey Tratner, Director Software Engineering, Bioinformatics at Myriad spoke at DNAnexus Connect, explaining how fast iteration works on the DNAnexus Platform. You can learn more by watching his talk or reading the summary below.
Typical pipeline development involves setting up an infrastructure, building a computation process, and analyzing the results. When adjustments are made, this process repeats as many times as necessary until the pipeline has been properly validated. With complex pipelines, this process can consume many resources and a lot of time. Myriad wanted a more efficient way to iterate on their pipelines so that they could optimize them faster. Fast R&D, as Myriad defines it, is characterized by an environment in which you can make adjustments easily, find answers quickly, and don’t have to think too much or second guess which areas of the pipeline you need to change when making adjustments.
Myriad reduced pipeline R&D from 2 weeks to 2 hours by leveraging tools that enable them to re-use computations.
The team at Myriad first demonstrated this concept when they performed a retrospective analysis with a new background normalization step, the tenth step of a 15-step workflow, on over 100,000 NIPT (non-invasive prenatal test) samples. Simply rerunning the entire modified workflow would have taken 2 weeks. Instead, Myriad reduced this time to two hours by rethinking the pipeline and leveraging tools that enable them to re-use computations.
Now codified, the approach in use at Myriad enables their team to make changes and iterate quickly, all with a focus on accuracy, reproducibility, and moving validated pipelines into production.
So how can you borrow from their approach and design your bioinformatics pipelines for faster iteration?
Make computational modules smaller
Although it’s tempting to use monorepositories when coding because they promote sharing, convenience, and low overhead, they don’t promote modularity within pipelines. And modularity is what enables you to scale quickly, re-use steps, and identify/debug problems. Myriad organized all the source code for workflows in monorepositories, but developed smart ways to break the code within them into smaller modules and only build the modules that have been modified.
Take advantage of tools that enable you to reuse computations
If you run an app with the same data and the same input files and parameters, your results should be equivalent. So if you are changing a step downstream, why run all of the steps that come before it if they’ve already been run? The DNAnexus Platform, for example, includes a Smart Reuse feature. This feature enables organizations to optionally reuse outputs of jobs that share the same executable and input IDs, even if these outputs are across projects. By reusing computational results, developers can dramatically speed the development of new workflows and reduce resources spent on testing at scale. To learn more about Smart Reuse visit our DNAnexus documentation here.
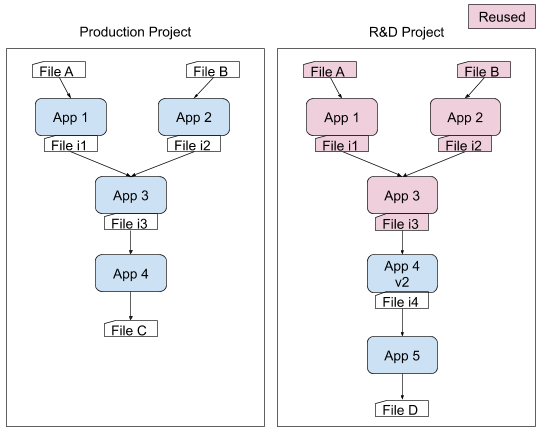
Use workflow tools to describe dependencies and manage the build process
Workflow tools, such as WDL (Workflow Description Language), make pipelines easier to express and build. With WDL, you can easily describe the module dependencies and track version changes to the workflow. It’s also very natural to integrate Docker with WDL, so if you’re using some sort of open-source container hub, you can simply edit one line in WDL and load a module of a different version with a new docker image. Myriad writes their bioinformatics pipelines in WDL and statically compiles them with dxWDL into DNAnexus workflows, streamlining the build process. Learn more about running Docker containers within DNAnexus apps or dxWDL from our DNAnexus documentation.

.png)
.png)
.png)