

On August 28, 2019, the Vertebrate Genome Project (VGP) announced the completion of 100 high-quality assembled, phased, and scaffolded genomes1. These new reference genomes include a wide variety of mammals, birds, reptiles, amphibians, and fish. The VGP is the multi-institutional and multinational collaboration that attempts to collect samples, develop methodologies, and craft the high-quality genomes of more than ten thousand species of vertebrates. In addition to species conservation, these genomes are irreplaceable resources to understand the biology of genomes and to study evolution at the genome-scale. The genome sequence is serving as a foundation of modern genetic study, and any defects in the assembled genomes could lead to incorrect biological conclusions. To ensure accuracy of data interpretation, the VGP established a new high standard in terms of continuity, completeness, and accuracy for these genomes. This standard requires over half of all assembled base-pairs to be included in contigs of greater than 1 million base pairs and half of all assembled base pairs to be included in scaffolds greater than 10 million base pairs. Additionally, 90% of base pairs must be assigned to a particular chromosome, the single base error rate must be less than 0.01%, and the genome needs to be phased and annotated. Given that none of the assembled genomes prior to this effort has this qualification, these are ambitious goals to meet on such a large scale.
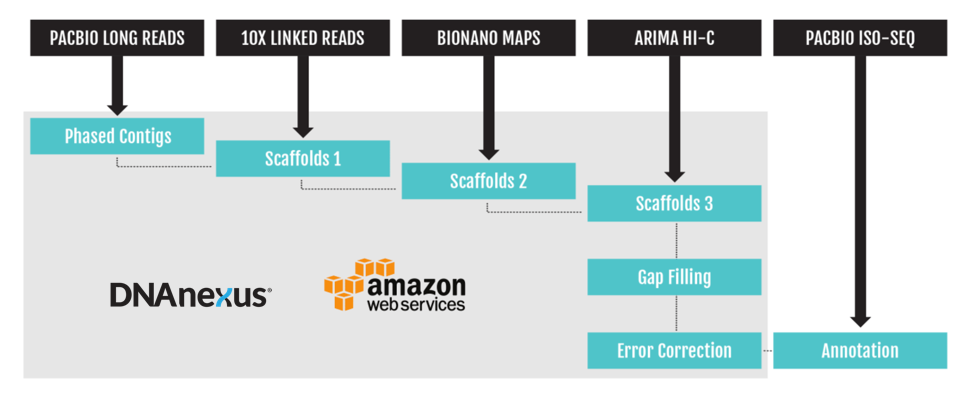
Constructing genomes at this unprecedented quality is very challenging. First, the computational requirement for genome assembly is extremely high. For example, a three gigabase mammalian genome needs roughly 30,000 core hours of computing with fluctuating memory and storage requirements to produce the final assembly. Such high computational loads would tax many local cluster environments when assembling a single organism, let alone assembling hundreds per year. Second, the bioinformatics pipeline for genome assembly and scaffolding is complicated and has to support multiple sequencing technologies. These pipelines are not trivial to set up and control the version across multiple institutions. Third, the existing genomic sequence for these species is very limited, therefore it is difficult to assess the accuracy of the assembly.
A cloud platform like the one offered by DNAnexus can help manage these challenges.
First, on the cloud, the computing resources can be deployed instantly and torn down after use. Additionally, different configurations of memory, storage, and computational cores can be deployed for each step, so that resource usage is optimized. Our scientists partnered with several lead scientists in this project including Adam Phillippy, Sergey Koren, and Arang Rhie from NIH, Olivier Fedrigo and Giulio Formenti from Rockefeller University, Richard Durbin, Shane McCarthy, and Kerstin Howe from Sanger Institute, Eugene Myers from Max-Planck-Gesellschaft, Erik Garrison from UCSC, and scientists from Pacific Biosciences to design these pipelines and optimize them to perform with high efficiency.
Second, the DNAnexus platform enables version control of the applications and workflows. This ensures that all participants are able to leverage the same tools configured in the same manner, and that the tools include all necessary dependencies. On a cloud platform like DNAnexus, the developer can encapsulate all the required programs and use the same version of those programs each time the application run.
Third, the data sharing feature of the cloud allows researchers across geographical locations to collaborate and inspect these genomes. For example, one lab may collect samples, another lab may perform the sequencing, and a third lab could perform the assembly. The data generated during this process can be coordinated and shared in a single DNAnexus project. Once the assembly process is complete, the assembly curation team from the Sanger institute plays a vital role in assembly generation by manually curating and correcting these genomes using various technologies and providing feedback on how to adjust the algorithms and pipelines to improve accuracy.
DNAnexus has been involved with the VGP since 2016. Our scientists, Brett Hannigan, Maria Simbirsky, and Arkarachai Fungtammasan, helped in constructing and maintaining the VGP assembly, phasing, scaffolding, and polishing pipelines on the cloud within the DNAnexus platform. The cloud is a suitable deployment system for highly fluctuating computing demand, version control, and collaboration, which address the challenges faced by VGP. Most of these assembled genomes generated by VGP have far exceeded the high standard set in this project.

The VGP group has an open door policy. We would like to encourage everyone who is interested to participate either in the form of donation, sample provider, or scientific collaboration. For more detail, visit https://vertebrategenomesproject.org/
Reference
1 https://phys.org/news/2019-08-vgp-largest-high-quality-genomes-iconic.html

.png)
.png)
.png)