At VIZBI 2018 (Visualizing Biological Data) [https://vizbi.org/2018/], I proposed a topic for a breakout session, called “One Genome Browser to Rule Them All”. This led to a very interesting discussion that I found fascinating and inspiring. I am writing this up here so we all have a place to continue the discussion and explore where to go next.
The basic idea is that there should be a genome browser that the whole community of researchers can use and build on, creating new track types as plugins that can be shared with other scientists, without the complexity of modifying the core framework.
We are at a point where web technologies have matured and made it possible to have more modularity and reusability of code on the web, just like what was possible in Java that enabled Cytoscape to have a very useful library of community-created plugins. This kind of large-scale collaboration to build an ideal genome browser is, I believe, at least an interesting thought experiment as the bioinformatics field matures and we are joined by many of the world’s biologists who rely increasingly on genome and transcriptome sequencing for their innovative research.
So here is roughly what we discussed during this session, with some blue sky wishful thinking:
Built for the web
We agreed that a JavaScript genome browser would be preferable to a desktop application given how much of genomics has moved onto the web with large databases of resources, cloud platforms, and lots of academic web applications that make genomics more accessible. If the genome browser is built for the web, each of these groups of services could have the genome browser embedded into it for better interactive exploration of the data. Among the genome browsers built for the web, some only need to be run from the front-end (IGV.js, pileup.js, Biodalliance) while others also need some back-end, server-side code or configuration to be used (JBrowse, NGB). It is significantly easier to embed the front-end browsers into a web application, whether that is a small academic project or a large platform like Galaxy or DNAnexus. If a back-end is the only way to improve performance, perhaps it can be an optional feature. I’m curious to hear people’s experiences on when having a back-end improves performance, and whether this is something that could be separated intelligently.
Future-proof
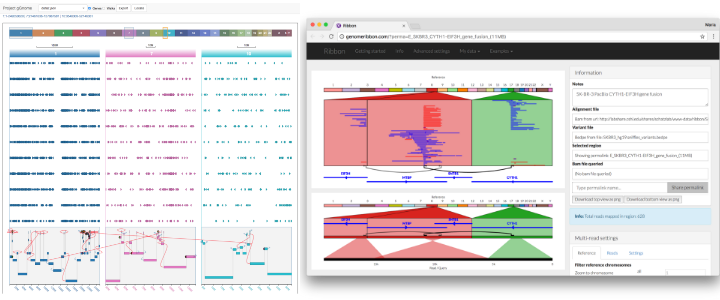
Importantly, this genome browser should also make fewer assumptions about how data will be visualized, to future-proof it against new technologies and ideas that need new types of visualization. Examples of limitations that are hard to overcome in current genome include the difficulty of plotting features that need to connect to multiple loci, which has become increasingly common in recent years for showing long reads (PacBio, Nanopore), linked reads (10X), and long-range variants including gene fusions, to name a few. Current genome browsers can sometimes show multiple loci next to each other, but the tracks cannot cut across and connect to more than one of these loci. See these examples of connecting across multiple loci from Ribbon and gGnome:
Ann Loraine, who is part of the IGB team, explained that the Integrated Genome Browser (IGB) is an older genome browser that predates IGV. The IGB team had attempted to add multi-locus functionality more recently, but they found that it was no longer possible to make such a fundamental change to the architecture of IGB. All software developers have to make assumptions when starting a project, but it’s important to examine those assumptions and consider what all the features are that we might eventually want to include. That is why future-proofing is such an important topic for this discussion.
To highlight another dimension of future-proofing, Valerie Schneider of the NCBI pointed out that a flood of new genomes, especially from the Vertebrate Genome Project, is one of the challenges she sees on the horizon for the NCBI’s own genome browser. Valerie also mentioned that enabling the genome browser connect to data from government and consortia resources would be very important.
Christian Stolte of the New York Genome Center showed a mini-browser within MetroNome that visually connects protein domains to their genomic coordinates along a gene. That example brought up an interesting point that non-genomic coordinates such as transcript and protein domains could also become first-class citizens. So then I guess we are no longer just talking about a genome browser but actually a multiome browser!
Notice that all of these use cases we discussed for future-proofing are already needed today and supported by niche mini-browsers. If you know of other current and predicted future use cases that are not already covered by existing genome browsers or by the ideas presented here, please add them in the comments, so we can all broaden our horizons and expand our vision for the ideal genome browser.
A library of community-contributed plugins
Before I built each of my own visualization tools, I tried to add the functionality I wanted to the genome browser I was using at the time, the desktop version of IGV. First, I needed to show two loci at the same time, with connecting lines between them for long-range variants. IGV could show two regions at once, but they were independent and I couldn’t draw lines across them — so I built a visualization tool called SplitThreader to show long-range variants as connecting lines between two loci. Later, I needed to show alignments of long reads in a way that you can tell where the alignments are along the read’s own length in addition to where they are on the reference, so I built Ribbon. The funny thing is, after building the parts of the visualization that are really novel, you still have to add several parts that normal genome browsers can do just fine, like drawing genes, variants, other features from a bed file. I would end up spending over 50% of my time implementing features that already exist elsewhere, and pretty shallow versions of them too.
Since starting my data visualization role at DNAnexus I also have a whole new appreciation for modularity because the platform is built to be able to run virtually any kind of analysis on any kind of data applicable in genomics. I dream of having a genome browser that could be equally flexible and modular, that anyone could contribute new types of visualization to, and that could be integrated and used anywhere from consortium database explorers to digital paper figures. If anyone can build an app on DNAnexus or Galaxy and publish it for others to use, then why is it not the same for adding a special track to a genome browser?
Conclusions
I would like to mention that the genome browsers out there right now are fantastic. When thinking about this ideal genome browser, there are several ideas that come directly from existing tools: IGV.js is very lightweight and does an excellent job of being embeddable anywhere, while JBrowse has a library of community-contributed plugins that I find very encouraging for collaboration in this field.
Genome browsers have significant similarities with each other, but each one also has its unique strengths that not all of them share. When I need to build a novel visualization for a particular data type, concept, or application, I would love to be able to just build a track into an already powerful and full-featured genome browser, instead of building the novel feature into its own tool and then adding genes, bed, and variants track types. The users of my tools would surely rather have the full power of an IGV or a JBrowse at their fingertips even while looking at their long-read alignments in a Ribbon browser.
Now I would love to hear what you think!
Are there any genome browser teams out there who have been thinking about modularity, plugins, future-proofing, and making a lightweight yet powerful genome browser for the web?



.png)
.png)
.png)