Test drive Sentieon’s pipelines on DNAnexus and see how you can achieve faster and more cost-efficient results with equivalent or improved accuracy and consistency
Editor’s Note: This blog post is written by Brendan Gallagher, Business Development Director at Sentieon.
We are excited to officially announce our collaboration with DNAnexus and the availability of our tools on the scalable cloud-based DNAnexus Platform. At Sentieon, we are enabling precision data for precision medicine by improving upon industry-leading bioinformatics tools. Our suite of secondary analysis tools produce equivalent or improved results while running much faster and more cost-efficient than GATK, MuTect and MuTect2 bioinformatics tools.
For a 2 month period, DNAnexus users will have license-free access to test, validate, and use:
- DNAseq (BWA + GATK 3.5) – From FASTQ / BAM to VCF
- TNseq (BWA + MuTect or MuTect2) – From FASTQ to VCF
- Rapid DNAseq (BWA + GATK 3.5) – From FASTQ to VCF in 1 hour
Register today to take advantage of this time-limited and license-free opportunity to access Sentieon pipelines on DNAnexus! General commercial availability begins April 8th.
Sentieon tools produce equivalent results to GATK/MuTect/MuTect2 suite of tools by using mathematical methods identical to the Broad Institute’s Best Practice Workflow pipeline. Sentieon has improved the efficiency of the computation algorithms and engineered robust software implementations to speed up the pipeline while providing equivalent or improved accuracy and 100% consistency.
Delivering Better Results
On the same compute infrastructure, Sentieon software is an order of magnitude faster in terms of core-hours, while producing 100% consistent results with no run-to-run difference. Sentieon software does not down-sample in high-coverage regions, enabling rigorous analysis for deep-coverage sequence data. By removing the down-sampling and other run-to-run error sources, Sentieon tools also improve the accuracy and quality of the results.
SPEED
Sentieon DNAseq processes a 30x NA12878 Genome from fastq to gVCF in ~6 hours and 15 minutes on a single 32 virtual core instance on the DNAnexus platform. Sentieon and DNAnexus are also offering a rapid-turnaround distributed version of the app that can complete a 30x genome in approximately 1 hour while adding minimal additional compute cost.
In our 2015 white paper comparing DNAseq to then-current GATK version 3.3, we showed that Sentieon DNAseq has a runtime improvement of 20-50x while producing identical results. Download the white paper.
The figure below shows the performance of our current version matching BWA-GATK3.5 on the DNAnexus Platform from FASTQ to VCF:
CONSISTENCY
Sentieon is 100% consistent and has no run-to-run variation You will get the same result every time you run an individual sample.
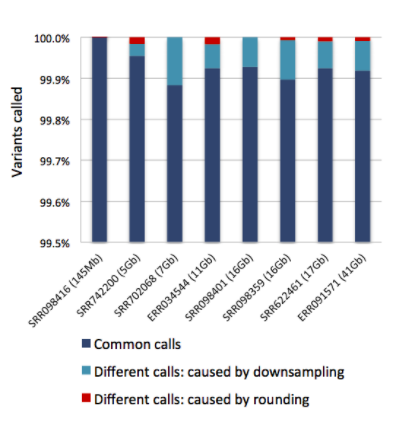
Sentieon Concordance to GATK – Identical within GATK’s run to run difference
The above figure shows the concordance analysis in our white paper. Over 99.8% of the variant calls produced by GATK 3.3 and Sentieon DNAseq were identical. After removing the differences from GATK downsampling, the variant calls were over 99.99% concordant. Learn more.
The same concordance performance is maintained in our current DNAseq version as compared to GATK3.5.
COST
With Sentieon, you can process a 30x genome from FASTQ to VCF for the previous price of an exome. DNAseq costs 5x less for whole exome sequencing (WES) and 7x less for whole genome sequencing (WGS) than GATK3 run on the DNAnexus Platform. For cost comparison purposes, please contact sales@dnanexus.com.
But don’t take my word for it.
Sentieon has been a top performer in many independent studies, and last year, we were recognized for accuracy and consistency in the precisionFDA community challenges: precisionFDA Consistency Challenge and precisionFDA Truth Challenge.
Furthermore, Sentieon software enables large cohort joint calls with tens of thousands of whole genomes without intermediate file merging, enabling much easier and much more efficient population-scale studies.
So go ahead, register here to try it out, and let us know what you think.
Email me anytime at brendan.gallagher@sentieon.com or talk to the DNAnexus team.
This is the start of a nice partnership as DNAnexus and Sentieon will continue to collaborate on the acceleration and improvement of genomic analysis by providing our customers with the most accurate and cost-conscious tools. We look forward to expanding the tools available on DNAnexus in the future.



.png)
.png)
.png)