Reference Genome Improvement, cfDNA for Transplantation, & de novo
Reconstruction of Familial Relationships
As the dust settles from American Society of Human Genetics 2015, we’d like to reflect and bring you some highlights from our lunchtime talk: Genomics and Informatics for the Next Five Years. We had a packed house and a stellar lineup of customers showcasing their cutting edge research on the DNAnexus Platform.
Tina Graves-Lindsay, Leader of the Reference Genomes Group at the McDonnell Genome Institute (MGI) at Washington University St. Louis, kicked off the session talking about the research involved in achieving the best human whole genome assembly. As a member of the Genome Reference Consortium, Tina’s team has been working to improve the current reference, GRCh38, and fixing a few genes that are not optimally represented for all individuals or ancestries.
The sequence plan starts with generating 60x coverage of PacBio long read data for a de novo assembly. From there, MGI incorporates BioNano or Dovetail data to create scaffolds that in some cases nearly cover entire chromosome arms. Since MGI is targeting difficult to assemble regions of the genome, they sequence bacterial artificial chromosomes (BACs) to fill the targeted regions and then incorporate all this data together to generate a very high quality whole genome assembly labeled the “Gold Genome”.
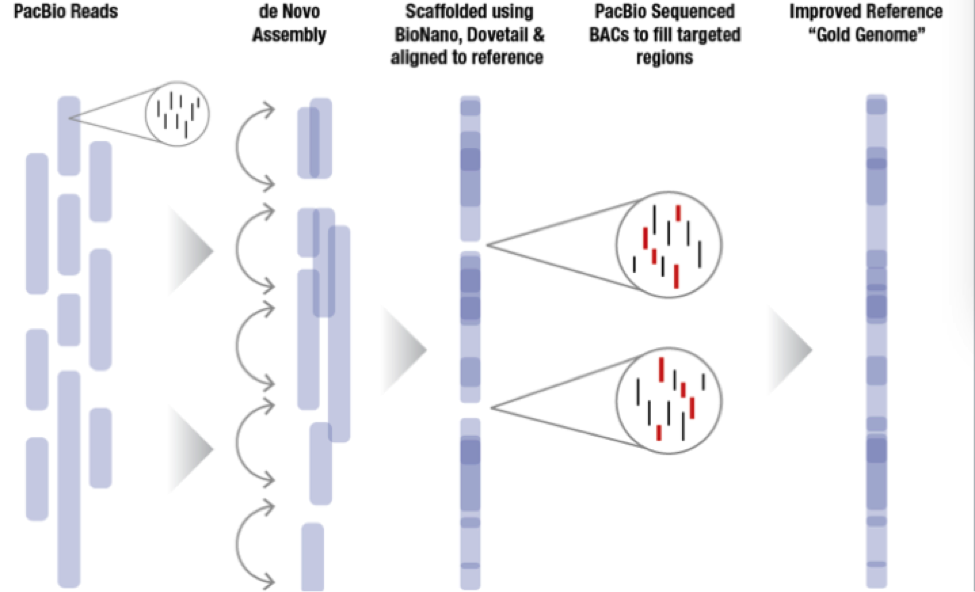
With these new technology improvements the MGI hopes to sequence more than 5 Gold Genomes as previously planned. With improvements in the BioNano pipelines, they hope to be able to automate some of the assembly breaks that need to be made to improve the final product. The DNAnexus Platform provides researchers with the computational power required and a turnkey solution for BioNano whole genome mapping and PacBio whole genome assembly. Labs of all sizes have fast and cost-effective access to the innovative technologies they need to reach their discoveries.
David Ross, Ph.D., Sr. Director of Bioinformatics at CareDx, presented his research on clinical-grade next generation sequencing (NGS) to monitor cell-free DNA (cfDNA) for transplantation. Post-transplant care is challenging. The main issue with organ transplantation is no longer organ rejection, but issues associated with over-immunosuppression; being able to balance immunosuppressive therapy with a titration tool is key.
Transplantation is the latest field to investigate the use of cfDNA for transplant surveillance. Research has shown that monitoring cfDNA for donor-specific homozygous single nucleotide polymorphisms (SNPs) can help identify transplant patients at risk of rejection. CareDx’s hypothesis is that cfDNA will be elevated at the time of rejection. CareDx’s cfDNA technology works by using NGS to measure donor-derived cfDNA (dd-cfDNA) as a percentage of total cfDNA. The dd-cfDNA can be detected at 0.25% level based on titration/calibration experiments; in contrast non-invasive prenatal testing cfDNA detection occurs between 5-20%.
CareDx is one of the few CLIA laboratories that has satisfied the rigorous review of the FDA when it received clearance for its laboratory developed AlloMap, a blood-based test used to monitor heart transplant recipients for rejection risk. CareDx worked closely with DNAnexus to complete the cloud-based computational analysis pipeline for the CareDx cfDNA test. The HIPAA compliant DNAnexus Platform, with full versioning and auditing, played a critical role in expediting CareDx’s cfDNA assay efforts, enabling them to deliver a novel surveillance management solution to the transplant setting.
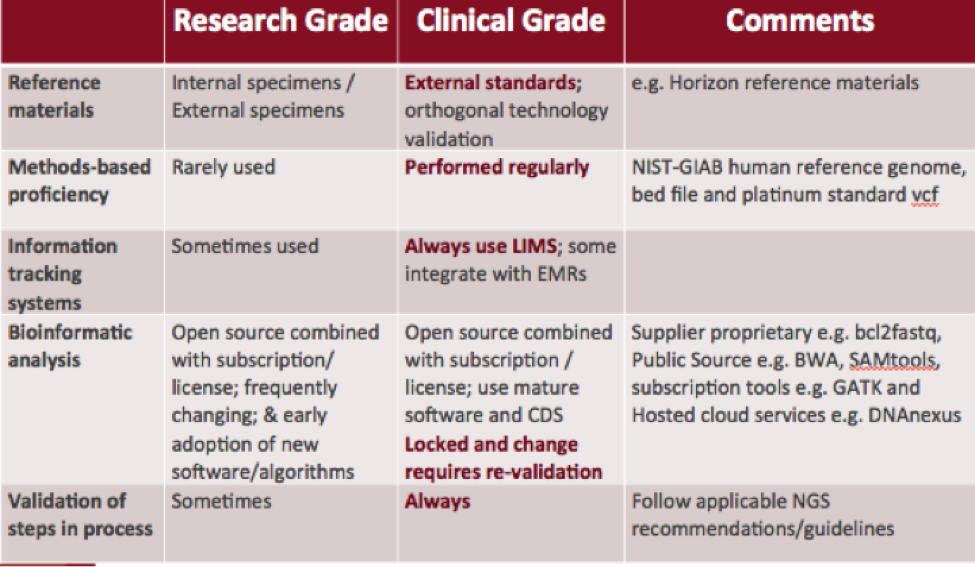
Research-Grade vs. Clinical-Grade NGS
Difference in methods, processes, and validation
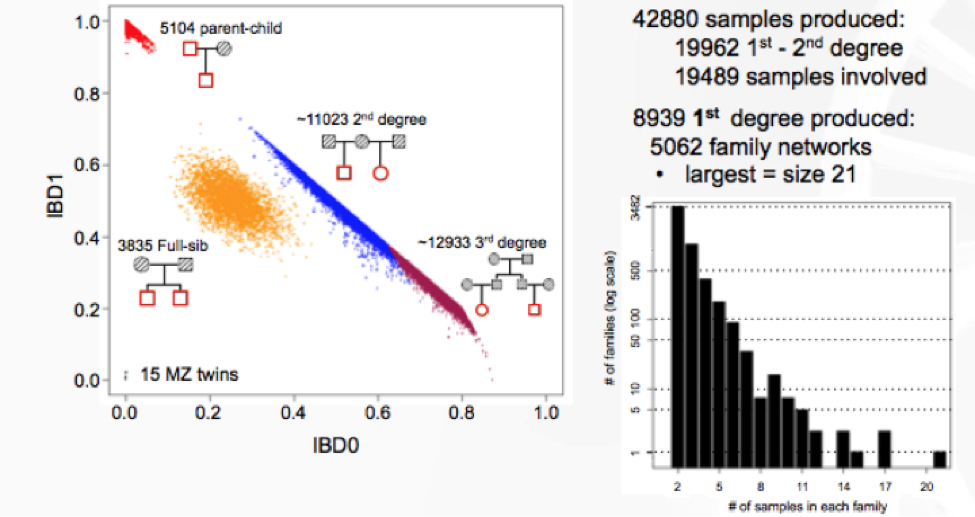
Lastly, Jeffrey Staples, Ph.D., Bioinformatics R&D Developer for Regeneron Genetics Center (RGC) discussed his latest research on de novo reconstruction of familial relationships in 42,880 exomes. The RGC has collaborated with Geisinger Healthcare System (GHS) to conduct large population-based studies integrating whole-exome sequencing (WES) with de-identified longitudinal health records. In this specific study, the RGC wanted to determine familial relationships in the GHS WES dataset, since this information can be used to build various studies and analyses requiring related or unrelated individuals. They used Identity By Decent (IBD), which measures a fraction of the genome shared between two people, as a measure of relatedness.
· IBD0 – proportion of the genome that is shared on 0 chromosomes
· IBD1 – proportion of the genome that is shared on 1 chromosomes
· IBD2 – proportion of the genome that is shared on 2 chromosomes
The results from distribution of IBD in >40,000 exomes showed 19,400 samples to have a first- or second-degree relatives in the dataset.
There is new thinking around handling relatedness in large-scale sequencing cohorts, performing de novo pedigree reconstruction to estimate the IBD between individuals and predict familial relationships. Initial findings show the Loss of Function (LOF) variant in CCR5, a protein on the surface of a white blood cell that is involved in the immune system, is transmitted through the family. The RGC is starting to leverage all family networks, including the 1,107-person second-degree family network.
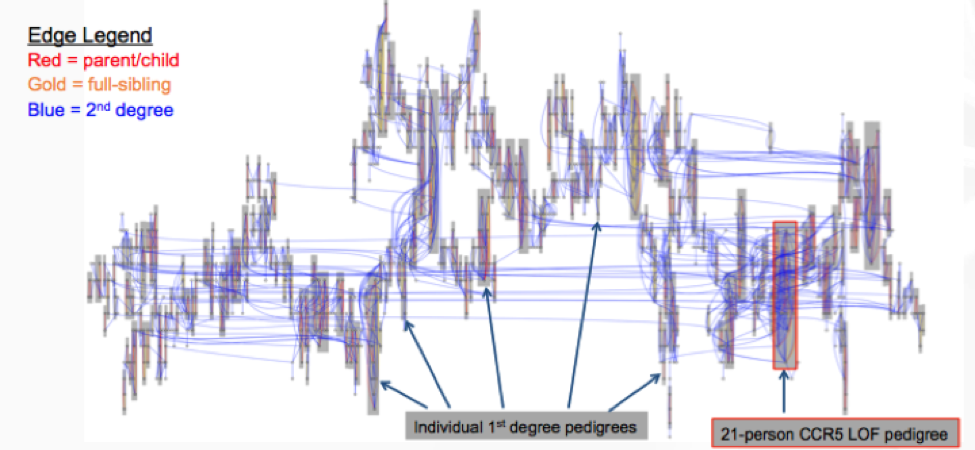
This massive relatedness is a big emerging opportunity for large scale sequencing studies and these pioneering methodologies will be invaluable to other large scale initiatives, leveraging families for discovery in precision medicine for all phenotypes going forward.
If you missed our lunchtime talk you can learn more about BioNano’s physical genome mapping integration with DNAnexus here, or visit DNAnexus.com to explore how we make it easier to manage your genomic data.

.png)
.png)
.png)