Today marks a major milestone for the ENCODE consortium! More than 30 papers will be published today in Nature, Genome Biology, and Genome Research from teams of scientists working on various facets of the project.
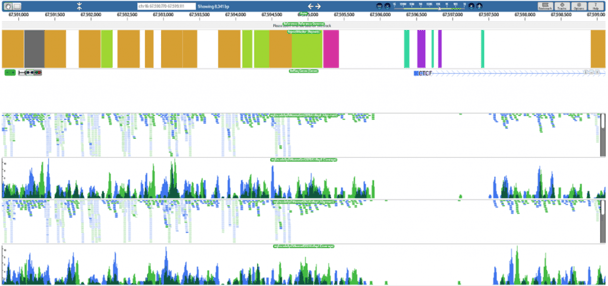
One of those, a publication in Genome Research, reports on a surprising level of heterogeneity among patterns of chromatin modifications as well as nucleosome positioning around regulatory elements such as transcription factor binding sites in the human genome. In the past, these genomic elements have been studied primarily by averaging patterns of chromatin marks across populations of sites, leading to the perception that patterns were much more uniform. The nucleosome positioning sequence data mapping and analysis was performed on the DNAnexus platform.
Lead author Anshul Kundaje was a postdoc for Serafim Batzoglou and Arend Sidow at Stanford University during the project reported in the paper. Now a research scientist at MIT, Kundaje says the work was an integral part of the ENCODE consortium’s efforts to elucidate functional elements in the human genome. The scientists looked at 119 human transcription factors and regulatory proteins to better understand how nucleosomes are positioned and how histone modifications are made around binding sites. In the paper, the authors report that asymmetry of nucleosome positioning and histone modifications is the rule, rather than the exception.
Kundaje and his colleagues relied on ChIP-seq data for the 119 transcription factors in a variety of cell types, with corresponding data for histone modifications. They also generated similar data for nucleosome positioning. To improve accuracy, the team sequenced extremely deeply, ultimately generating some 5 billion reads on the SOLiD sequencing platform. “The data sets were incredibly massive,” Kundaje says. “Processing these data sets locally was quite a challenge.” The group turned to DNAnexus, uploading their sequence files to the cloud and preprocessing the data with the company’s probabilistic mapping tool. “DNAnexus made that process incredibly simple,” he adds.
Using a new tool they developed — the Clustered AGgregation Tool (CAGT) for pattern discovery — the scientists found that nucleosome positioning and histone modification at transcription factor binding sites is far more diverse than was previously thought. Rather than averaging across the regions as most studies have done, the new clustering tool was able to analyze the differences in magnitude, shapes, and orientation of the many patterns identified.
“What we found is that the results you get from the clustering approach are dramatically different from what you get by simply averaging across all types,” Kundaje says. “We found a large diversity of patterns of histone modifications as well as nucleosome positioning around almost every transcription factor binding site.”
Even the well-known and remarkably well studied transcription factor CTCF, long established as an insulator, was found to have surrounding chromatin patterns pointing to other functions throughout the genome.
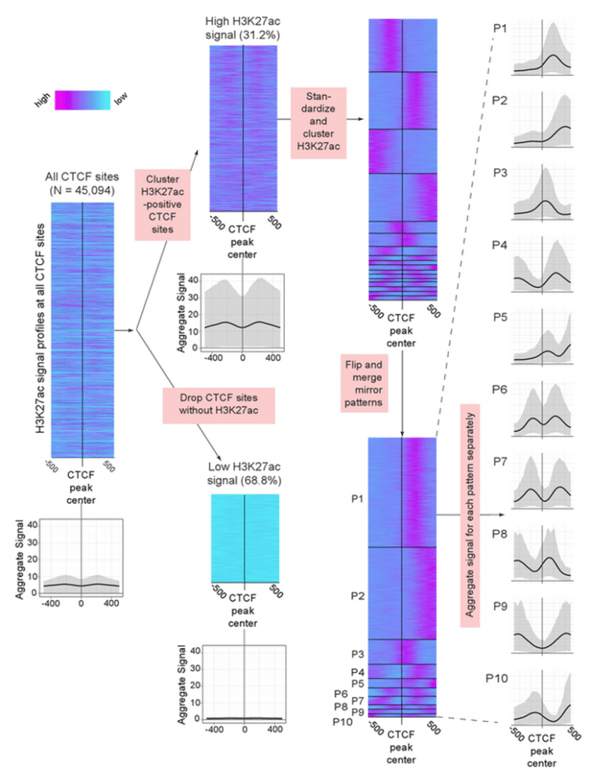
The authors used their clustering tool to group the patterns into some 25 distinct signatures “that completely capture the diversity of all the modifications across all binding sites in a variety of cell types,” Kundaje says. The method uses ‘metapatterns’ to explain that diversity, and that information can reveal the function of these elements in context. “By accounting for combinatorial relationships between various binding events and how they affect chromatin, this gives you a more complete biological sense of what a transcription factor is doing in a cell type,” he adds.
Kundaje is already following up on this study by looking at other species to see whether the heterogeneity of modification patterns holds true in other organisms. He continues to use DNAnexus for analysis of sequencing data, especially in read mapping, quality control, and genome browsing, he says.
Using DNAnexus for the team’s ENCODE study “made the process significantly easier,” Kundaje adds, noting that the cloud provider’s direct integration of the genome browser was particularly helpful. DNAnexus allowed Kundaje and his colleagues to go from data to visualization with minimal processing steps in between, he says. “It frees up your time to focus on the more interesting work.”
For a glimpse of some of Kundaje’s data, DNAnexus has made the 20 samples available on DNAnexus in the Public Data folder, called Encode. Click here to sign up for a free account.

.png)
.png)
.png)