This week, we and our collaborators at the Human Genome Sequencing Center at Baylor College of Medicine uploaded a preprint for Parliament2, an optimized method to deploy multiple of structural variant callers and combine their outputs into a single consensus set of structural variants. The code for Parliament2 is open source on Github, available as a Docker container, and as an app on DNAnexus. In this blog post, we explore the reasons to run Parliament2 and add structural variant calling to your genomic analyses.
Introduction
Structural variants are large (50 bp+) rearrangements in a genome. While SNPs and Indels are much smaller than the size of an individual Illumina read (~150 bp) and can be observed directly, structural variants are large and complex enough that mapping in and around them is difficult. Thus, structural variants must be indirectly inferred by the “shadow” they cast in the genome.
Structural variants come in a number of categories: deletions, insertions, inversions, translocations, duplications, mobile elements, and repeat expansions/contractions. The signals used to detect them are similarly diverse, including read depth, split-read mapping, read-pair orientation, insert-size distribution, and soft-clipping of sequences. These signals can vary strongly depending on the conditions and design of the sequencing experiment.
Method authors exploit different combinations of these signals to find structural variants. Some of the many methods include Breakdancer, Breakseq2, CNVnator, Delly, Lumpy, and Manta to name a few. We have developed Parliament2 as a method that allows a user to quickly and efficiently run multiple methods in a single execution and to combine them in a manner that maximizes discovery power, and does so quickly, taking 3 hours of wall-clock time and around 60 core-hours to run.
Parliament2 represents a re-engineering of the first version of Parliament, which was developed in a collaboration between the HGSC and DNAnexus. The first version of Parliament could incorporate both long-read and short-read data and used assembly to validate events. Parliament2 currently only works on Illumina data and has been designed to achieve significant improvements in speed and efficiency, allowing its application at the scale of 100,000+ WGS cohorts. Parliament1 remains available in the DNAnexus app library.
Because the Parliament2 manuscript goes into significant detail, in this blog, we will summarize the key scientific and logistic advantages of Parliament2.
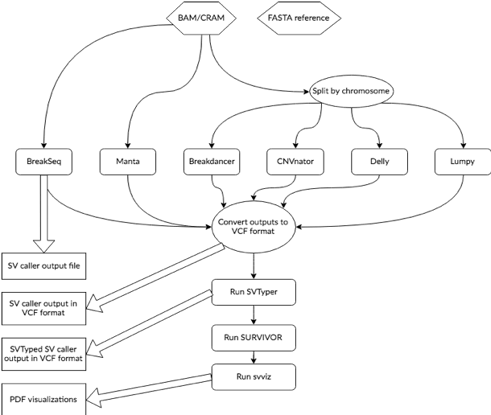
Parliament2 Runs Multiple Structural Variant Callers, Combines Them in a Standard Format, and Genotypes Them
The figure to the right shows a schematic for the data flow and tools used in Parliament2. Starting from a BAM or CRAM and its associated reference genome file, Parliament2 runs any combination of the callers Breakdancer, Breakseq2, CNVnator, Delly, Lumpy, and Manta as specified by the user.
Calls are integrated with SURVIVOR. This step combines events discovered in multiple methods, preserving which methods support each event. These calls are genotyped with SVTyper, which confirms events with an orthogonal computational method, resulting in higher precision and a genotype call.
Parliament2 is available as a Docker image, which comes pre-installed with all of the required dependencies for each program (important since some of them have mutual incompatibilites in libraries like pysam and therefore require their own environment). Parliament2 can be executed from any environment with a docker pull -> docker run command and a user can select which set of programs to run. For example, if you only want to run CNVnator, Parliament2 provides an easy way to do so.
Parliament2 Uses Multiple Parallelization Strategies to be Faster and More Cost-Efficient
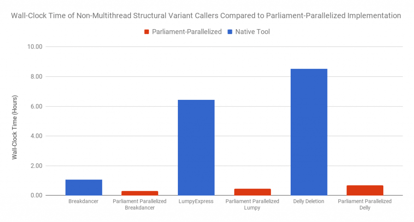 Breakdancer, CNVnator, Delly, and Lumpy do not effectively multi-thread (though Lumpy has been improved as smoove for this purpose). By managing parallel execution and result merging across chromosomes, Parliament2 can achieve significant speed increases compared to the most naive use of these tools.
Breakdancer, CNVnator, Delly, and Lumpy do not effectively multi-thread (though Lumpy has been improved as smoove for this purpose). By managing parallel execution and result merging across chromosomes, Parliament2 can achieve significant speed increases compared to the most naive use of these tools.
This improvement speeds up individual tools. The larger advantage for compute resources comes from parallel execution. Each of the tools used has different requirements for CPU, disk I/O, and RAM. Running multiple methods together at the same time smooths the resource draw.
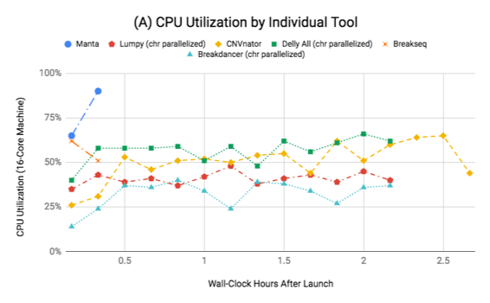 The chart (A) to the right shows the percent core utilization achieved on a 16-core machine when running any of methods individually. Manta and Breakseq both finish quickly, while the other methods complete over the time of 2-3 hours, each using a fraction of total machine resources. Remember that this generally leads to wasted resources, as you are typically paying for a full machine’s worth of time regardless of how much you use.
The chart (A) to the right shows the percent core utilization achieved on a 16-core machine when running any of methods individually. Manta and Breakseq both finish quickly, while the other methods complete over the time of 2-3 hours, each using a fraction of total machine resources. Remember that this generally leads to wasted resources, as you are typically paying for a full machine’s worth of time regardless of how much you use.
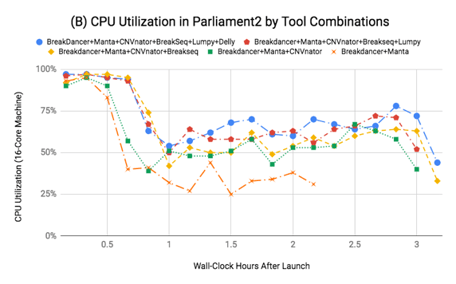 The chart to the left shows the CPU utilization in Parliament2 when running various combinations of callers. Overall, the machine is far more efficiently utilized by running multiple methods concurrently. From running Breakdancer, Manta, and CNVnator, with only 10 more minutes of job time, you can add Lumpy, Delly, and Breakseq almost “for free”.
The chart to the left shows the CPU utilization in Parliament2 when running various combinations of callers. Overall, the machine is far more efficiently utilized by running multiple methods concurrently. From running Breakdancer, Manta, and CNVnator, with only 10 more minutes of job time, you can add Lumpy, Delly, and Breakseq almost “for free”.
Precision and Recall of Parliament2 (and Individual Methods)
In the remaining sections, we will analyze the precision of Parliament2 as well as that of the individual tools that compose Parliament. These comparisons are based on the Genome in a Bottle Structural Variant v0.6 Truth Set using the Truvari tool by Adam English of Spiral Genetics. Because the SV discovery power of short reads is weak for inserts in all methods (maximum recall of 17%), we will mostly present the deletion benchmarks here.
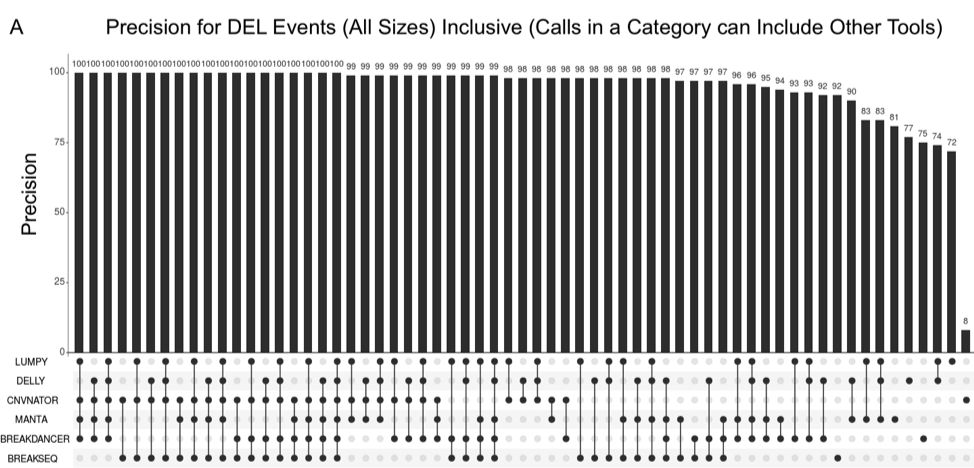
Parliament2 Achieves High Precision with Combinations of Calls
The plot above is an UpSet plot, which shows how the combination of callers that supports an event reflects the probability that a call is correct. A filled black circle indicates a category where a tool called an event, so in the column where Breakdancer, Manta, and Breakseq each have a filled black circle, it means AT LEAST these methods made a call. This chart indicates that Manta achieves the best precision of individual methods at 90%, but calls supported by multiple methods can reach very high precision (99%-100%).
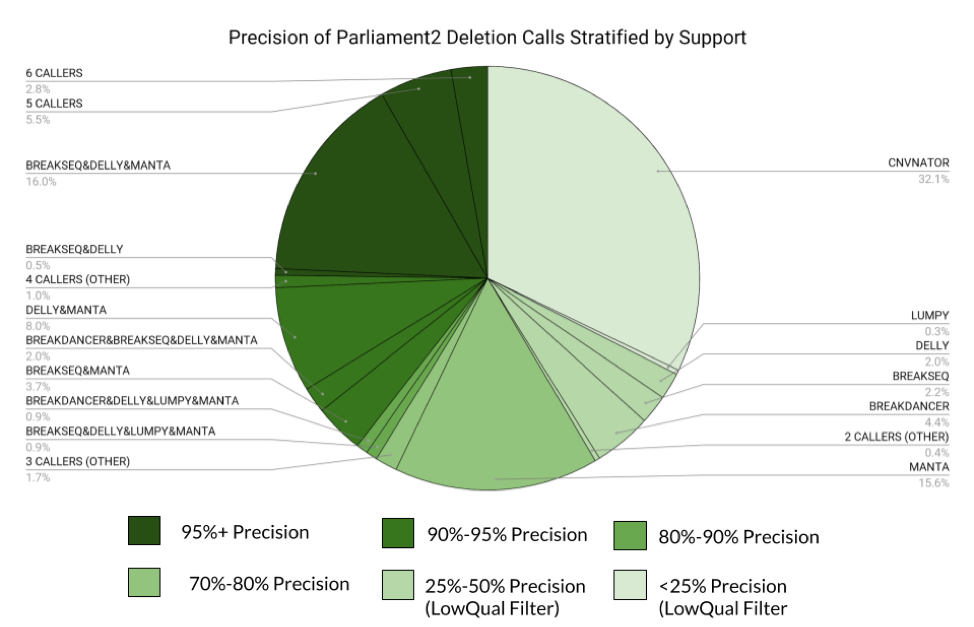
The figure above shows a breakdown of all calls made by Parliament2 with their category of support. This gives a feel for how many calls are made at each confidence level. Any category below 50% precision is given a LowQual filter field in the VCF.
Parliament2 Gives Confidence Scores Based on Size and Event Type
In addition to filtering calls in the VCF, this same concept allows us to give a Phred-encoded quality value to each call based on the size of the event and the type of event. This allows a downstream analyst to create their own filtering criteria based on quality. 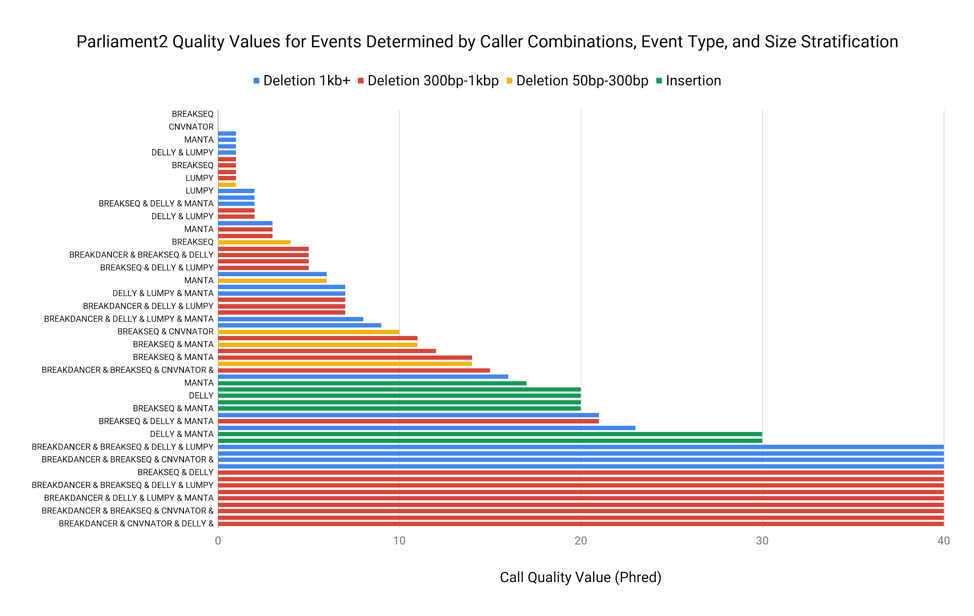
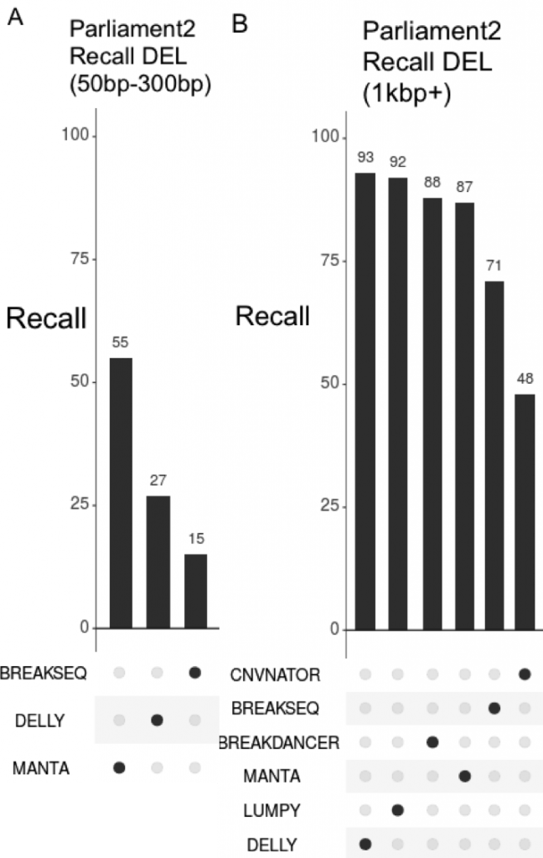
Parliament2 Discovers More Events by Combining the Strengths of Multiple Methods
By running more tools, Parliament2 can find events that individual methods might miss. In particular, this is driven by differences in discovery power between different size ranges.
Only certain methods call deletions less than 300bp, and among those that do so, Manta demonstrates by far the highest recall at 55%.
However, for events larger than 1kbp, Manta is the fourth-weakest method.
In general, smaller SV events are more common in humans, meaning that the aggregate accuracy numbers are disproportionately influenced by small events.
Having additional callers to supplement the strong overall performance of Manta allows for additional discovery power, particularly for larger events. This can be especially important since large events disrupt greater fractions of the genome and may be disproportionately more likely to drive genetic disease.
In Conclusion
It is natural to fixate on questions of who has “the best” method. The Parliament methods (1&2) are not about writing better individual tools; the concept relies on finding the right way to ensure that in the combination of many methods, the whole is more than the sum of its parts.
Parliament illustrates a fact that can seem counter-intuitive. Even in the extreme situation where you have tool A which can discover every event that tool B finds and with fewer false positives, you can still do better by running both and gaining knowledge about which events are supported by both as opposed to only one.
Many scientists have brought an enormous range of opinions, effort, and creativity to bear in the field of calling structural variation. Parliament is a very specific and tangible manifestation of a statement which is even more true for science as a whole: even the most experienced can gain from the wisdom of others. And we are stronger working together than we are apart.
By fortunate chance, Samantha was presenting her work at 23andMe Genome Day when Tony Blair happened to be visiting. Samantha had the opportunity to present Parliament2 to the former UK Prime Minister. When learning about how Parliament enables the combination of many opinions for a greater outcome, Mr. Blair said something to the effect of “I hope that this is so”.


.png)
.png)
.png)