DNAnexus was featured in a couple of posters at AGBT this year. DNAnexus scientist, Andrew Carroll, was in the poster hall, sharing data from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium genomic analysis project. In addition, Narayanan Veeraraghavan, Lead Programmer Scientist at Baylor’s Human Genome Sequencing Center, presented a poster focused their own variant-calling pipeline, Mercury that was built on the cloud via the DNAnexus platform.
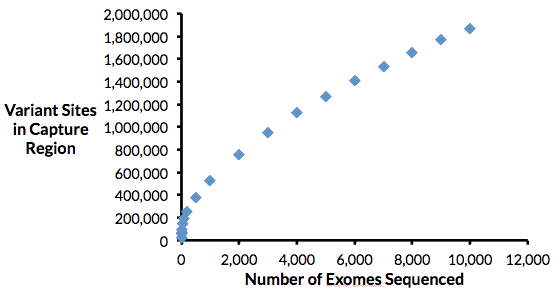
Andrew presented never seen before data that was generated by Baylor’s Human Genome Sequencing Center using DNAnexus to run Mercury, its own exome and genome analysis pipeline. Interestingly, it was found as exome numbers increase, so do new variants found. Here’s a graph demonstrating that each new exome analyzed yielded novel genetic variation, rather than reaching a plateau after some critical mass of exomes:
For Andrew’s perspective on the poster, check out this short video:
The heart of Narayanan’s poster centered around Mercury on the Cloud Analytics (MOCA), an automated NGS pipeline HGSC developed, which upon being ported to DNAnexus was used successfully for novel gene discovery in two large-scale projects: CHARGE and Baylor-Hopkins Center for Mendelian Genomics (BHCMG). Some of the key features in this production pipeline cited in the poster include: zero setup, on-demand scale-up, version control, reproducibility, compliance with CAP/CLIA/HIPAA, and ISO 27001 data handling and security and compliance.
We look forward to seeing alternate workflows being built on DNAnexus by CHARGE commons to benchmark studies without the need to install or configure analysis tools.

.png)
.png)
.png)